The Murine Tcl1 Oncogene: Embryonic and Lymphoid Cell Expression
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
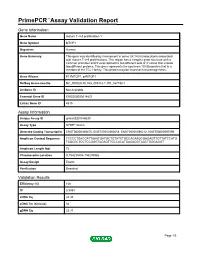
Primepcr™Assay Validation Report
PrimePCR™Assay Validation Report Gene Information Gene Name mature T-cell proliferation 1 Gene Symbol MTCP1 Organism Human Gene Summary This gene was identified by involvement in some t(X;14) translocations associated with mature T-cell proliferations. This region has a complex gene structure with a common promoter and 5' exon spliced to two different sets of 3' exons that encode two different proteins. This gene represents the upstream 13 kDa protein that is a member of the TCL1 family. This protein may be involved in leukemogenesis. Gene Aliases P13MTCP1, p8MTCP1 RefSeq Accession No. NC_000023.10, NG_005114.1, NT_167198.1 UniGene ID Not Available Ensembl Gene ID ENSG00000214827 Entrez Gene ID 4515 Assay Information Unique Assay ID qHsaCED0048630 Assay Type SYBR® Green Detected Coding Transcript(s) ENST00000369476, ENST00000362018, ENST00000596212, ENST00000597096 Amplicon Context Sequence TCCCCTGACCATTAAATGATGCTGTATCTGCCACAAGCGAGAGTTGTTATCCATG TAGCGCTCCTCCGGGTAGAGTTGCCACATGAGAGGTAGCTGGGAGGT Amplicon Length (bp) 72 Chromosome Location X:154293804-154293986 Assay Design Exonic Purification Desalted Validation Results Efficiency (%) 106 R2 0.9994 cDNA Cq 24.34 cDNA Tm (Celsius) 82 gDNA Cq 25.37 Page 1/5 PrimePCR™Assay Validation Report Specificity (%) 100 Information to assist with data interpretation is provided at the end of this report. Page 2/5 PrimePCR™Assay Validation Report MTCP1, Human Amplification Plot Amplification of cDNA generated from 25 ng of universal reference RNA Melt Peak Melt curve analysis of above amplification Standard -

Structural Studies of the Complex Between Akt-In and the Akt2-PH Domain Suggest That the Peptide Acts As an Allosteric Inhibitor of the Akt Kinase
The Open Spectroscopy Journal, 2009, 3, 65-76 65 Open Access Structural Studies of the Complex Between Akt-in and the Akt2-PH Domain Suggest that the Peptide Acts as an Allosteric Inhibitor of the Akt Kinase Virginie Ropars1,2,3, Philippe Barthe1,2,3, Chi-Shien Wang4, Wenlung Chen4, Der-Lii M. Tzou4,5, Anne Descours6, Loïc Martin6, Masayuki Noguchi7, Daniel Auguin1,2,3,,¶ and Christian Roumestand*,1,2,3 1CNRS UMR 5048, Centre de Biochimie Structurale, Montpellier, France 2INSERM U554, Montpellier, France 3Université Montpellier I et II, Montpellier, France 4Department of Applied Chemistry, National Chiayi University, Chiayi 60004, Taiwan, ROC 5Institute of Chemistry, Academia Sinica, Nankang, Taipei 11529, Taiwan, ROC 6CEA, iBiTecs, Service d’Ingénierie Moléculaire des Protéines, 91191 Gif sur Yvette, France 7Division of Cancer Biology, Institute for Genetic Medicine, Hokkaido University, N15 W7, Kita-ku, Sapporo 060-0815, Japan Abstract: Serine/threonine kinase Akt plays a central role in the regulation of cell survival and proliferation. Hence, the search for Akt specific inhibitors constitutes an attractive strategy for anticancer therapy. We have previously demonstrated that the proto-oncogene TCL1 coactivates Akt upon binding to its Plekstrin Homology Domain, and we proposed a model for the structure of the complex TCL1:Akt2-PHD. This model led to the rational design of Akt-in, a peptide inhibitor spanning the A ß-strand of human TCL1 that binds Akt2 PH domain and inhibits the kinase activation. In the present report, we used NMR spectroscopy to determine the 3D structure of the peptide free in solution and bound to Akt2-PHD. -

Association of Gene Ontology Categories with Decay Rate for Hepg2 Experiments These Tables Show Details for All Gene Ontology Categories
Supplementary Table 1: Association of Gene Ontology Categories with Decay Rate for HepG2 Experiments These tables show details for all Gene Ontology categories. Inferences for manual classification scheme shown at the bottom. Those categories used in Figure 1A are highlighted in bold. Standard Deviations are shown in parentheses. P-values less than 1E-20 are indicated with a "0". Rate r (hour^-1) Half-life < 2hr. Decay % GO Number Category Name Probe Sets Group Non-Group Distribution p-value In-Group Non-Group Representation p-value GO:0006350 transcription 1523 0.221 (0.009) 0.127 (0.002) FASTER 0 13.1 (0.4) 4.5 (0.1) OVER 0 GO:0006351 transcription, DNA-dependent 1498 0.220 (0.009) 0.127 (0.002) FASTER 0 13.0 (0.4) 4.5 (0.1) OVER 0 GO:0006355 regulation of transcription, DNA-dependent 1163 0.230 (0.011) 0.128 (0.002) FASTER 5.00E-21 14.2 (0.5) 4.6 (0.1) OVER 0 GO:0006366 transcription from Pol II promoter 845 0.225 (0.012) 0.130 (0.002) FASTER 1.88E-14 13.0 (0.5) 4.8 (0.1) OVER 0 GO:0006139 nucleobase, nucleoside, nucleotide and nucleic acid metabolism3004 0.173 (0.006) 0.127 (0.002) FASTER 1.28E-12 8.4 (0.2) 4.5 (0.1) OVER 0 GO:0006357 regulation of transcription from Pol II promoter 487 0.231 (0.016) 0.132 (0.002) FASTER 6.05E-10 13.5 (0.6) 4.9 (0.1) OVER 0 GO:0008283 cell proliferation 625 0.189 (0.014) 0.132 (0.002) FASTER 1.95E-05 10.1 (0.6) 5.0 (0.1) OVER 1.50E-20 GO:0006513 monoubiquitination 36 0.305 (0.049) 0.134 (0.002) FASTER 2.69E-04 25.4 (4.4) 5.1 (0.1) OVER 2.04E-06 GO:0007050 cell cycle arrest 57 0.311 (0.054) 0.133 (0.002) -

Regulation of the Akt Kinase by Interacting Proteins
Oncogene (2005) 24, 7401–7409 & 2005 Nature Publishing Group All rights reserved 0950-9232/05 $30.00 www.nature.com/onc Regulation of the Akt kinase by interacting proteins Keyong Du1 and Philip N Tsichlis*,1 1Molecular Oncology Research Institute, Tufts-New England Medical Center, Boston, MA 02111, USA Ten years ago, it was observed that the Akt kinase revolutionized our understanding of cell function at is activated by phosphorylation via a phosphoinositide the molecular level.These technologies also allowed 3-kinase (PI-3K)-dependent process. This discovery gene- the identification of a large array of protein–protein rated enormous interest because it provided a link between interaction motifs that facilitate our understanding of PI-3K, an enzyme known to play a critical role in cellular signal transduction by providing clues on the potential physiology, and its downstream targets. Subsequently, it function of novel signaling proteins (Barnouin, 2004; was shown that the activity of the core components of the Miller and Stagljar, 2004). ‘PI-3K/Akt pathway’ is modulated by a complex network Akt1, also known as protein kinase Ba (PKBa) of regulatory proteins and pathways. Some of the Akt- (Bellacosa et al., 1991; Coffer and Woodgett, 1991; binding partners modulate its activation by external Jones et al., 1991), is the founding member of a protein signals by interacting with different domains of the Akt kinase family composed of three members, Akt1, Akt2, protein. This review focuses on the Akt interacting and Akt3.Akt family members regulate a diverse proteins and the mechanisms by which they regulate Akt array of cellular functions, including apoptosis, cellular activation. -

Tcl1 Enhances Akt Kinase Activity and Mediates Its Nuclear Translocation
Tcl1 enhances Akt kinase activity and mediates its nuclear translocation Yuri Pekarsky*, Anatoliy Koval*, Cora Hallas*, Roberta Bichi*, Maria Tresini*, Scott Malstrom*, Giandomenico Russo†, Philip Tsichlis*, and Carlo M. Croce*‡ *Kimmel Cancer Center, Jefferson Medical College, Philadelphia, PA 19107; and †Istituto Dermopatico dell’Immacolata, Laboratory of Vascular Pathology, 00167 Rome, Italy Contributed by Carlo M. Croce, December 20, 1999 The TCL1 oncogene at 14q32.1 is involved in the development of wortmannin, a (PI-3K)-kinase inhibitor, completely inhibits the human mature T-cell leukemia. The mechanism of action of Tcl1 is activation of Akt (reviewed in ref. 9). Recent studies showed that unknown. Because the virus containing the v-akt oncogene causes Akt is a key player in the transduction of antiapoptotic and T-cell lymphoma in mice and Akt is a key player in transduction of proliferative signals in T-cells (refs. 10 and 11; reviewed in ref. antiapoptotic and proliferative signals in T-cells, we investigated 9). Activated Akt enhances both cell cycle progression and IL2 whether Akt and Tcl1 function in the same pathway. Coimmuno- production through the inhibition by phosphorylation of the precipitation experiments showed that endogenous Akt1 and Tcl1 proapoptotic factor Bad (11). Introduction of a constitutively physically interact in the T-cell leukemia cell line SupT11; both activated AKT1 transgene under the control of the proximal lck proteins also interact when cotransfected into 293 cells. Using promoter causes T-cell lymphomas in mice (S.M. and P.T., several AKT1 constructs in cotransfection experiments, we deter- unpublished data). In cultured cells, Akt1 can be localized in mined that this interaction occurs through the pleckstrin homology both the nucleus and cytoplasm (12). -

NRF1) Coordinates Changes in the Transcriptional and Chromatin Landscape Affecting Development and Progression of Invasive Breast Cancer
Florida International University FIU Digital Commons FIU Electronic Theses and Dissertations University Graduate School 11-7-2018 Decipher Mechanisms by which Nuclear Respiratory Factor One (NRF1) Coordinates Changes in the Transcriptional and Chromatin Landscape Affecting Development and Progression of Invasive Breast Cancer Jairo Ramos [email protected] Follow this and additional works at: https://digitalcommons.fiu.edu/etd Part of the Clinical Epidemiology Commons Recommended Citation Ramos, Jairo, "Decipher Mechanisms by which Nuclear Respiratory Factor One (NRF1) Coordinates Changes in the Transcriptional and Chromatin Landscape Affecting Development and Progression of Invasive Breast Cancer" (2018). FIU Electronic Theses and Dissertations. 3872. https://digitalcommons.fiu.edu/etd/3872 This work is brought to you for free and open access by the University Graduate School at FIU Digital Commons. It has been accepted for inclusion in FIU Electronic Theses and Dissertations by an authorized administrator of FIU Digital Commons. For more information, please contact [email protected]. FLORIDA INTERNATIONAL UNIVERSITY Miami, Florida DECIPHER MECHANISMS BY WHICH NUCLEAR RESPIRATORY FACTOR ONE (NRF1) COORDINATES CHANGES IN THE TRANSCRIPTIONAL AND CHROMATIN LANDSCAPE AFFECTING DEVELOPMENT AND PROGRESSION OF INVASIVE BREAST CANCER A dissertation submitted in partial fulfillment of the requirements for the degree of DOCTOR OF PHILOSOPHY in PUBLIC HEALTH by Jairo Ramos 2018 To: Dean Tomás R. Guilarte Robert Stempel College of Public Health and Social Work This dissertation, Written by Jairo Ramos, and entitled Decipher Mechanisms by Which Nuclear Respiratory Factor One (NRF1) Coordinates Changes in the Transcriptional and Chromatin Landscape Affecting Development and Progression of Invasive Breast Cancer, having been approved in respect to style and intellectual content, is referred to you for judgment. -

The Human Gene Connectome As a Map of Short Cuts for Morbid Allele Discovery
The human gene connectome as a map of short cuts for morbid allele discovery Yuval Itana,1, Shen-Ying Zhanga,b, Guillaume Vogta,b, Avinash Abhyankara, Melina Hermana, Patrick Nitschkec, Dror Friedd, Lluis Quintana-Murcie, Laurent Abela,b, and Jean-Laurent Casanovaa,b,f aSt. Giles Laboratory of Human Genetics of Infectious Diseases, Rockefeller Branch, The Rockefeller University, New York, NY 10065; bLaboratory of Human Genetics of Infectious Diseases, Necker Branch, Paris Descartes University, Institut National de la Santé et de la Recherche Médicale U980, Necker Medical School, 75015 Paris, France; cPlateforme Bioinformatique, Université Paris Descartes, 75116 Paris, France; dDepartment of Computer Science, Ben-Gurion University of the Negev, Beer-Sheva 84105, Israel; eUnit of Human Evolutionary Genetics, Centre National de la Recherche Scientifique, Unité de Recherche Associée 3012, Institut Pasteur, F-75015 Paris, France; and fPediatric Immunology-Hematology Unit, Necker Hospital for Sick Children, 75015 Paris, France Edited* by Bruce Beutler, University of Texas Southwestern Medical Center, Dallas, TX, and approved February 15, 2013 (received for review October 19, 2012) High-throughput genomic data reveal thousands of gene variants to detect a single mutated gene, with the other polymorphic genes per patient, and it is often difficult to determine which of these being of less interest. This goes some way to explaining why, variants underlies disease in a given individual. However, at the despite the abundance of NGS data, the discovery of disease- population level, there may be some degree of phenotypic homo- causing alleles from such data remains somewhat limited. geneity, with alterations of specific physiological pathways under- We developed the human gene connectome (HGC) to over- come this problem. -

Estudo Da Deficiência Mental De Herança Ligada Ao Cromossomo X
JOSÉ OLIVEIRA DOS SANTOS ESTUDO DA DEFICIÊNCIA MENTAL DE HERANÇA LIGADA AO CROMOSSOMO X (Versão Corrigida) Tese apresentada ao Instituto de Biociências da Universidade de São Paulo, para a obtenção de Título de Doutor em Ciências, na Área de Biologia/Genética São Paulo 2013 Orientadora: Angela M. Vianna Morgante OLIVEIRA DOS SANTOS, JOSÉ ESTUDO DA DEFICIÊNCIA MENTAL DE HERANÇA LIGADA AO CROMOSSOMO X Tese (Doutorado) - Instituto de Biociências da Universidade de São Paulo, Departamento de Genética e Biologia Evolutiva 1. Deficiência Mental 2. Deficiência Mental com Herança Ligada ao X 3. Microrrearranjos Cromossômicos 4. Inativação do Cromossomo X Universidade de São Paulo. Instituto de Biociências. Departamento de Genética e Biologia Evolutiva COMISSÃO JULGADORA _____________________________ ____________________________ _____________________________ ____________________________ __________________________________ Orientadora Este trabalho foi realizado com auxílio financeiro da FAPESP (Fundação de Amparo à Pesquisa do Estado de São Paulo) concedido à orientadora (FAPESP-CEPID 98/14254- 2) e bolsa da CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior) concedida ao aluno (CAPES-DS). AGRADECIMENTOS Os meus sinceros agradecimentos a todos que de alguma forma contribuíram para a realização deste trabalho. Ao Departamento de Genética e Biologia Evolutiva do Instituto e Biociências da Universidade de São Paulo, pela infraestrutura que permitiu a realização deste trabalho. À Dra. Angela M. Vianna-Morgante, pela orientação neste projeto, por permitir que eu fizesse parte deste excelente grupo de pesquisa, pelos ensinamentos e pela confiança depositada em mim. Ao Dr. Paulo Alberto Otto, pelos ensinamentos, pelo auxílio nas análises e por estar sempre disposto a ajudar no que fosse necessário. O exame clínico que realizou nos pacientes foi fundamental para este trabalho. -

UC San Diego Electronic Theses and Dissertations
UC San Diego UC San Diego Electronic Theses and Dissertations Title Cardiac Stretch-Induced Transcriptomic Changes are Axis-Dependent Permalink https://escholarship.org/uc/item/7m04f0b0 Author Buchholz, Kyle Stephen Publication Date 2016 Peer reviewed|Thesis/dissertation eScholarship.org Powered by the California Digital Library University of California UNIVERSITY OF CALIFORNIA, SAN DIEGO Cardiac Stretch-Induced Transcriptomic Changes are Axis-Dependent A dissertation submitted in partial satisfaction of the requirements for the degree Doctor of Philosophy in Bioengineering by Kyle Stephen Buchholz Committee in Charge: Professor Jeffrey Omens, Chair Professor Andrew McCulloch, Co-Chair Professor Ju Chen Professor Karen Christman Professor Robert Ross Professor Alexander Zambon 2016 Copyright Kyle Stephen Buchholz, 2016 All rights reserved Signature Page The Dissertation of Kyle Stephen Buchholz is approved and it is acceptable in quality and form for publication on microfilm and electronically: Co-Chair Chair University of California, San Diego 2016 iii Dedication To my beautiful wife, Rhia. iv Table of Contents Signature Page ................................................................................................................... iii Dedication .......................................................................................................................... iv Table of Contents ................................................................................................................ v List of Figures ................................................................................................................... -

The Int22h1/Int22h2-Mediated Xq28 Duplication Syndrome: an Intersection Between Neurodevelopment, Immunology, and Cancer
G C A T T A C G G C A T genes Review The int22h1/int22h2-Mediated Xq28 Duplication Syndrome: An Intersection between Neurodevelopment, Immunology, and Cancer Rami A. Ballout 1,* and Ayman W. El-Hattab 2,3,* 1 Lipoprotein Metabolism Section, Translational Vascular Medicine Branch, National Heart, Lung and Blood Institute (NHLBI), National Institutes of Health (NIH), Bethesda, MD 20814, USA 2 Department of Clinical Sciences, College of Medicine, University of Sharjah, Sharjah P. O. Box 27272, United Arab Emirates 3 Clinical Genetics, University Hospital Sharjah, Sharjah P. O. Box 72772, United Arab Emirates * Correspondence: [email protected] (R.A.B.); [email protected] (A.W.E.-H.); Tel.: +1-(202)-417-5832 (R.A.B.); +97-15-0887-5123 (A.W.E.-H.) Abstract: The int22h1/int22h2-mediated Xq28 duplication syndrome is a rare X-linked intellectual disability syndrome (XLIDS) arising from a duplication of the segment between intron 22 homologous regions 1 and 2, on the q28 subregion of the X chromosome. The main clinical features of the syndrome include intellectual disability, neurobehavioral abnormalities, and dysmorphic facial features. Due to the X-linked nature of the syndrome, affected males exhibit more severe phenotypes compared with heterozygous females. A unique distinguishing feature of the syndrome across the sexes, however, is a peculiar combination of recurrent sinopulmonary infections and atopy exclusively seen in a subset of affected males. In addition to the ‘typical’ 0.5 Mb duplication detected in most cases reported to date with the syndrome, a shortened centromeric version, and another 0.2 Mb telomerically shifted Citation: Ballout, R.A.; El-Hattab, A.W. -

Monoclonal Antibody to TCL1A / TCL1
AM26558BT-N OriGene Technologies Inc. OriGene EU Acris Antibodies GmbH 9620 Medical Center Drive, Ste 200 Schillerstr. 5 Rockville, MD 20850 32052 Herford UNITED STATES GERMANY Phone: +1-888-267-4436 Phone: +49-5221-34606-0 Fax: +1-301-340-8606 Fax: +49-5221-34606-11 [email protected] [email protected] Monoclonal Antibody to TCL1A / TCL1 - Biotin Alternate names: T-cell leukemia/lymphoma protein 1A, TCL-1 Catalog No.: AM26558BT-N Quantity: 0.1 ml Concentration: 0.5 mg/ml Background: TCL1 (T cell leukemia/lymphoma 1), MTCP1 (mature T cell proliferation 1) and TCL1b belong to the TCL1 proto-oncogene family. The TCL1 gene at chromosome 14q32.1 is co mmonly activated in T cell neoplasms by chromosome r earrangements such as inversions inv(14)(q11;q32) and translocations t(14;14)(q11;q32) or t(7;14)(q35;q32). TCL1 expression is limited to immature thymocytes in early T-cell progenitors and from pre-B to mature B cells in the B-cell lineage. The TCL1/MTCP1/TCL1b proto-oncogene activation is the hallmark of human T-cell prolymphocytic leukemia (T-PLL), a form of adult leukemia. In addition to T- PLL, TCL1 is overexpressed in Burkitt's lymphoma cell lines, the majority of AIDS-related non-Hodgkin’s lymphoma-designated immunoblastic lymphoma plasmacytoids, lymphoblastic lymphoma, chronic lymphocytic leukemia, mantle cell lymphoma, follicular lymphoma, diffuse large B-cell lymphoma, and primary cutaneous B-cell lymphoma. It is also implicated in the development of hematopoietic abnormalities in patients with ataxia- telangiectasia. The members of the TCL1 proto-oncogene family bind to the serine/threonine kinase Akt (PKB), increasing its phosphorylation status and kinase activity. -
Structural Characterization of the Human Proteome
Article Structural Characterization of the Human Proteome Arne Mu¨ller,1 Robert M. MacCallum,1,4 and Michael J.E. Sternberg1,2,3 1Biomolecular Modelling Laboratory, Cancer Research UK, London, United Kingdom; 2Department of Biological Sciences, Structural Bioinformatics Group, Imperial College of Science, Technology and Medicine, South Kensington, London, United Kingdom This paper reports an analysis of the encoded proteins (the proteome) of the genomes of human, fly, worm, yeast, and representatives of bacteria and archaea in terms of the three-dimensional structures of their globular domains together with a general sequence-based study. We show that 39% of the human proteome can be assigned to known structures. We estimate that for 77% of the proteome, there is some functional annotation, but only 26% of the proteome can be assigned to standard sequence motifs that characterize function. Of the human protein sequences, 13% are transmembrane proteins, but only 3% of the residues in the proteome form membrane-spanning regions. There are substantial differences in the composition of globular domains of transmembrane proteins between the proteomes we have analyzed. Commonly occurring structural superfamilies are identified within the proteome. The frequencies of these superfamilies enable us to estimate that 98% of the human proteome evolved by domain duplication, with four of the 10 most duplicated superfamilies specific for multicellular organisms. The zinc-finger superfamily is massively duplicated in human compared to fly and worm, and occurrence of domains in repeats is more common in metazoa than in single cellular organisms. Structural superfamilies over- and underrepresented in human disease genes have been identified.