Gene Expression Profiles Alteration After Infection of Virus, Bacteria, And
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
Primepcr™Assay Validation Report
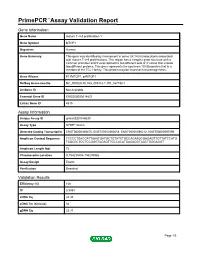
PrimePCR™Assay Validation Report Gene Information Gene Name mature T-cell proliferation 1 Gene Symbol MTCP1 Organism Human Gene Summary This gene was identified by involvement in some t(X;14) translocations associated with mature T-cell proliferations. This region has a complex gene structure with a common promoter and 5' exon spliced to two different sets of 3' exons that encode two different proteins. This gene represents the upstream 13 kDa protein that is a member of the TCL1 family. This protein may be involved in leukemogenesis. Gene Aliases P13MTCP1, p8MTCP1 RefSeq Accession No. NC_000023.10, NG_005114.1, NT_167198.1 UniGene ID Not Available Ensembl Gene ID ENSG00000214827 Entrez Gene ID 4515 Assay Information Unique Assay ID qHsaCED0048630 Assay Type SYBR® Green Detected Coding Transcript(s) ENST00000369476, ENST00000362018, ENST00000596212, ENST00000597096 Amplicon Context Sequence TCCCCTGACCATTAAATGATGCTGTATCTGCCACAAGCGAGAGTTGTTATCCATG TAGCGCTCCTCCGGGTAGAGTTGCCACATGAGAGGTAGCTGGGAGGT Amplicon Length (bp) 72 Chromosome Location X:154293804-154293986 Assay Design Exonic Purification Desalted Validation Results Efficiency (%) 106 R2 0.9994 cDNA Cq 24.34 cDNA Tm (Celsius) 82 gDNA Cq 25.37 Page 1/5 PrimePCR™Assay Validation Report Specificity (%) 100 Information to assist with data interpretation is provided at the end of this report. Page 2/5 PrimePCR™Assay Validation Report MTCP1, Human Amplification Plot Amplification of cDNA generated from 25 ng of universal reference RNA Melt Peak Melt curve analysis of above amplification Standard -

Structural Studies of the Complex Between Akt-In and the Akt2-PH Domain Suggest That the Peptide Acts As an Allosteric Inhibitor of the Akt Kinase
The Open Spectroscopy Journal, 2009, 3, 65-76 65 Open Access Structural Studies of the Complex Between Akt-in and the Akt2-PH Domain Suggest that the Peptide Acts as an Allosteric Inhibitor of the Akt Kinase Virginie Ropars1,2,3, Philippe Barthe1,2,3, Chi-Shien Wang4, Wenlung Chen4, Der-Lii M. Tzou4,5, Anne Descours6, Loïc Martin6, Masayuki Noguchi7, Daniel Auguin1,2,3,,¶ and Christian Roumestand*,1,2,3 1CNRS UMR 5048, Centre de Biochimie Structurale, Montpellier, France 2INSERM U554, Montpellier, France 3Université Montpellier I et II, Montpellier, France 4Department of Applied Chemistry, National Chiayi University, Chiayi 60004, Taiwan, ROC 5Institute of Chemistry, Academia Sinica, Nankang, Taipei 11529, Taiwan, ROC 6CEA, iBiTecs, Service d’Ingénierie Moléculaire des Protéines, 91191 Gif sur Yvette, France 7Division of Cancer Biology, Institute for Genetic Medicine, Hokkaido University, N15 W7, Kita-ku, Sapporo 060-0815, Japan Abstract: Serine/threonine kinase Akt plays a central role in the regulation of cell survival and proliferation. Hence, the search for Akt specific inhibitors constitutes an attractive strategy for anticancer therapy. We have previously demonstrated that the proto-oncogene TCL1 coactivates Akt upon binding to its Plekstrin Homology Domain, and we proposed a model for the structure of the complex TCL1:Akt2-PHD. This model led to the rational design of Akt-in, a peptide inhibitor spanning the A ß-strand of human TCL1 that binds Akt2 PH domain and inhibits the kinase activation. In the present report, we used NMR spectroscopy to determine the 3D structure of the peptide free in solution and bound to Akt2-PHD. -

Association of Gene Ontology Categories with Decay Rate for Hepg2 Experiments These Tables Show Details for All Gene Ontology Categories
Supplementary Table 1: Association of Gene Ontology Categories with Decay Rate for HepG2 Experiments These tables show details for all Gene Ontology categories. Inferences for manual classification scheme shown at the bottom. Those categories used in Figure 1A are highlighted in bold. Standard Deviations are shown in parentheses. P-values less than 1E-20 are indicated with a "0". Rate r (hour^-1) Half-life < 2hr. Decay % GO Number Category Name Probe Sets Group Non-Group Distribution p-value In-Group Non-Group Representation p-value GO:0006350 transcription 1523 0.221 (0.009) 0.127 (0.002) FASTER 0 13.1 (0.4) 4.5 (0.1) OVER 0 GO:0006351 transcription, DNA-dependent 1498 0.220 (0.009) 0.127 (0.002) FASTER 0 13.0 (0.4) 4.5 (0.1) OVER 0 GO:0006355 regulation of transcription, DNA-dependent 1163 0.230 (0.011) 0.128 (0.002) FASTER 5.00E-21 14.2 (0.5) 4.6 (0.1) OVER 0 GO:0006366 transcription from Pol II promoter 845 0.225 (0.012) 0.130 (0.002) FASTER 1.88E-14 13.0 (0.5) 4.8 (0.1) OVER 0 GO:0006139 nucleobase, nucleoside, nucleotide and nucleic acid metabolism3004 0.173 (0.006) 0.127 (0.002) FASTER 1.28E-12 8.4 (0.2) 4.5 (0.1) OVER 0 GO:0006357 regulation of transcription from Pol II promoter 487 0.231 (0.016) 0.132 (0.002) FASTER 6.05E-10 13.5 (0.6) 4.9 (0.1) OVER 0 GO:0008283 cell proliferation 625 0.189 (0.014) 0.132 (0.002) FASTER 1.95E-05 10.1 (0.6) 5.0 (0.1) OVER 1.50E-20 GO:0006513 monoubiquitination 36 0.305 (0.049) 0.134 (0.002) FASTER 2.69E-04 25.4 (4.4) 5.1 (0.1) OVER 2.04E-06 GO:0007050 cell cycle arrest 57 0.311 (0.054) 0.133 (0.002) -

Survival of Miamiensis Avidus (Ciliophora: Scuticociliatia) from Antibody-Dependent Complement Killing
www.ksfp.org 한국어병학회지 제28권 제3호 (2015) pISSN 1226-0819, eISSN 2233-5412 J. Fish Pathol., 28(3) : 171~174 http://dx.doi.org/10.7847/jfp.2015.28.3.171 Note Survival of Miamiensis avidus (Ciliophora: Scuticociliatia) from antibody-dependent complement killing Eun Hye Lee1, Yue Jai Kang2 and Ki Hong Kim3† 1Imported Food Analysis Division, Ministry of Food and Drug Safety, Busan Regional Office, Busan 48562, South Korea 2Department of Aquatic Life and Medical Sciences, Sun Moon University, Asan-si, Chungnam, 31460, South Korea 3Department of Aquatic Life Medicine, Pukyong National University, Busan 48513, South Korea Previously, we had reported that some Miamiensis avidus, a major pathogen of scuticociliatosis in cultured olive flounder, strongly agglutinated by flounder immune sera could escape from the agglutinated mass within a few hours. In the present study, we observed that M. avidus not only escaped from the agglutinated mass but also conducted division(s) before shedding its old covering. Furthermore, ciliates that survived the antibody-dependent complement killing (ADCK) assay were not killed even when re-exposed to a freshly prepared ADCK assay. This result suggests that the liberated ciliates from the ADCK assay might change not only their i-antigen types but also the epitopes of major surface antigens, which debilitate antibody-mediated complement killing ability. Key words: Miamiensis avidus, Agglutination, Antibody-dependent complement killing, Division, Survival A protein called immobilization antigen (i-antigen) is a facultative parasitic ciliate and has been a culprit is known as the major protein covering ciliates sur- of mass mortalities in cultured marine fish, such as face including cilia. -

Regulation of the Akt Kinase by Interacting Proteins
Oncogene (2005) 24, 7401–7409 & 2005 Nature Publishing Group All rights reserved 0950-9232/05 $30.00 www.nature.com/onc Regulation of the Akt kinase by interacting proteins Keyong Du1 and Philip N Tsichlis*,1 1Molecular Oncology Research Institute, Tufts-New England Medical Center, Boston, MA 02111, USA Ten years ago, it was observed that the Akt kinase revolutionized our understanding of cell function at is activated by phosphorylation via a phosphoinositide the molecular level.These technologies also allowed 3-kinase (PI-3K)-dependent process. This discovery gene- the identification of a large array of protein–protein rated enormous interest because it provided a link between interaction motifs that facilitate our understanding of PI-3K, an enzyme known to play a critical role in cellular signal transduction by providing clues on the potential physiology, and its downstream targets. Subsequently, it function of novel signaling proteins (Barnouin, 2004; was shown that the activity of the core components of the Miller and Stagljar, 2004). ‘PI-3K/Akt pathway’ is modulated by a complex network Akt1, also known as protein kinase Ba (PKBa) of regulatory proteins and pathways. Some of the Akt- (Bellacosa et al., 1991; Coffer and Woodgett, 1991; binding partners modulate its activation by external Jones et al., 1991), is the founding member of a protein signals by interacting with different domains of the Akt kinase family composed of three members, Akt1, Akt2, protein. This review focuses on the Akt interacting and Akt3.Akt family members regulate a diverse proteins and the mechanisms by which they regulate Akt array of cellular functions, including apoptosis, cellular activation. -

Disease of Aquatic Organisms 86:163
Vol. 86: 163–167, 2009 DISEASES OF AQUATIC ORGANISMS Published September 23 doi: 10.3354/dao02113 Dis Aquat Org NOTE DNA identification of ciliates associated with disease outbreaks in a New Zealand marine fish hatchery 1, 1 1 1 2 P. J. Smith *, S. M. McVeagh , D. Hulston , S. A. Anderson , Y. Gublin 1National Institute of Water and Atmospheric Research (NIWA), Private Bag 14901, Wellington, New Zealand 2NIWA, Station Road, Ruakaka, Northland 0166, New Zealand ABSTRACT: Ciliates associated with fish mortalities in a New Zealand hatchery were identified by DNA sequencing of the small subunit ribosomal RNA gene (SSU rRNA). Tissue samples were taken from lesions and gill tissues on freshly dead juvenile groper, brain tissue from adult kingfish, and from ciliate cultures and rotifers derived from fish mortality events between January 2007 and March 2009. Different mortality events were characterized by either of 2 ciliate species, Uronema marinum and Miamiensis avidus. A third ciliate, Mesanophrys carcini, was identified in rotifers used as food for fish larvae. Sequencing part of the SSU rRNA provided a rapid tool for the identification and mon- itoring of scuticociliates in the hatchery and allowed the first identification of these species in farmed fish in New Zealand. KEY WORDS: Small subunit ribosomal RNA gene · Scuticociliatosis · Uronema marinum · Miamiensis avidus · Mesanophrys carcini · Groper · Polyprion oxygeneios · Kingfish · Seriola lalandi Resale or republication not permitted without written consent of the publisher INTRODUCTION of ciliate pathogens in fin-fish farms (Kim et al. 2004a,b, Jung et al. 2007) and in crustacea (Ragan et The scuticociliates are major pathogens in marine al. -

Tcl1 Enhances Akt Kinase Activity and Mediates Its Nuclear Translocation
Tcl1 enhances Akt kinase activity and mediates its nuclear translocation Yuri Pekarsky*, Anatoliy Koval*, Cora Hallas*, Roberta Bichi*, Maria Tresini*, Scott Malstrom*, Giandomenico Russo†, Philip Tsichlis*, and Carlo M. Croce*‡ *Kimmel Cancer Center, Jefferson Medical College, Philadelphia, PA 19107; and †Istituto Dermopatico dell’Immacolata, Laboratory of Vascular Pathology, 00167 Rome, Italy Contributed by Carlo M. Croce, December 20, 1999 The TCL1 oncogene at 14q32.1 is involved in the development of wortmannin, a (PI-3K)-kinase inhibitor, completely inhibits the human mature T-cell leukemia. The mechanism of action of Tcl1 is activation of Akt (reviewed in ref. 9). Recent studies showed that unknown. Because the virus containing the v-akt oncogene causes Akt is a key player in the transduction of antiapoptotic and T-cell lymphoma in mice and Akt is a key player in transduction of proliferative signals in T-cells (refs. 10 and 11; reviewed in ref. antiapoptotic and proliferative signals in T-cells, we investigated 9). Activated Akt enhances both cell cycle progression and IL2 whether Akt and Tcl1 function in the same pathway. Coimmuno- production through the inhibition by phosphorylation of the precipitation experiments showed that endogenous Akt1 and Tcl1 proapoptotic factor Bad (11). Introduction of a constitutively physically interact in the T-cell leukemia cell line SupT11; both activated AKT1 transgene under the control of the proximal lck proteins also interact when cotransfected into 293 cells. Using promoter causes T-cell lymphomas in mice (S.M. and P.T., several AKT1 constructs in cotransfection experiments, we deter- unpublished data). In cultured cells, Akt1 can be localized in mined that this interaction occurs through the pleckstrin homology both the nucleus and cytoplasm (12). -

NRF1) Coordinates Changes in the Transcriptional and Chromatin Landscape Affecting Development and Progression of Invasive Breast Cancer
Florida International University FIU Digital Commons FIU Electronic Theses and Dissertations University Graduate School 11-7-2018 Decipher Mechanisms by which Nuclear Respiratory Factor One (NRF1) Coordinates Changes in the Transcriptional and Chromatin Landscape Affecting Development and Progression of Invasive Breast Cancer Jairo Ramos [email protected] Follow this and additional works at: https://digitalcommons.fiu.edu/etd Part of the Clinical Epidemiology Commons Recommended Citation Ramos, Jairo, "Decipher Mechanisms by which Nuclear Respiratory Factor One (NRF1) Coordinates Changes in the Transcriptional and Chromatin Landscape Affecting Development and Progression of Invasive Breast Cancer" (2018). FIU Electronic Theses and Dissertations. 3872. https://digitalcommons.fiu.edu/etd/3872 This work is brought to you for free and open access by the University Graduate School at FIU Digital Commons. It has been accepted for inclusion in FIU Electronic Theses and Dissertations by an authorized administrator of FIU Digital Commons. For more information, please contact [email protected]. FLORIDA INTERNATIONAL UNIVERSITY Miami, Florida DECIPHER MECHANISMS BY WHICH NUCLEAR RESPIRATORY FACTOR ONE (NRF1) COORDINATES CHANGES IN THE TRANSCRIPTIONAL AND CHROMATIN LANDSCAPE AFFECTING DEVELOPMENT AND PROGRESSION OF INVASIVE BREAST CANCER A dissertation submitted in partial fulfillment of the requirements for the degree of DOCTOR OF PHILOSOPHY in PUBLIC HEALTH by Jairo Ramos 2018 To: Dean Tomás R. Guilarte Robert Stempel College of Public Health and Social Work This dissertation, Written by Jairo Ramos, and entitled Decipher Mechanisms by Which Nuclear Respiratory Factor One (NRF1) Coordinates Changes in the Transcriptional and Chromatin Landscape Affecting Development and Progression of Invasive Breast Cancer, having been approved in respect to style and intellectual content, is referred to you for judgment. -

California Ground Squirrel
all 2017 f The Newsletter of the Hayward Shoreline Interpretive Center Volume 32, Number 4 A facility of Biodiversity in the Bay Hayward B y D o m i ni c i n n Area Recreation & Park District recently traveled with my family to Van- California.” I remember asking my advisor couver, in Canada’s British Columbia, if I could take a replacement class because Iwhich got me thinking about why we love studying plants sounded very hard and (to visiting new places so much. Reasons vary, be honest) a little boring. My advisor con- UPCOMING EVENTS from expanding knowledge, visiting other vinced me to take the class by reminding AT THE SHORELINE cultures, finding new challenges, getting me that if I wanted to study animals, I’d SEPTEMBER away from life’s business, or snapping Insta- have to understand the plants that make • Sleep with the Fishes: gram pictures. I love traveling. I often find habitats suitable for them. I ended up lov- Family Sleepover Night myself planning make-believe trips to the Sat. Sep. 23, 6:00pm-10:00am Great Barrier Reef in Australia, rainforests in South America, or deserts in Africa. My OCTOBER not only is California h desire is to explore natural areas I’ve never Birding:a Murmur Has It ia beautiful, it’s also one • y w n been to, encounter plants and animals I Sat. Oct. 28, 11:00am-2:00pma o r r d, c a lif can’t see at home in the San Francisco Bay of the most biodiverse NOVEMBER Area, and fish for species not found in areas in the world • Tidepooling Time California’s waters. -

The Human Gene Connectome As a Map of Short Cuts for Morbid Allele Discovery
The human gene connectome as a map of short cuts for morbid allele discovery Yuval Itana,1, Shen-Ying Zhanga,b, Guillaume Vogta,b, Avinash Abhyankara, Melina Hermana, Patrick Nitschkec, Dror Friedd, Lluis Quintana-Murcie, Laurent Abela,b, and Jean-Laurent Casanovaa,b,f aSt. Giles Laboratory of Human Genetics of Infectious Diseases, Rockefeller Branch, The Rockefeller University, New York, NY 10065; bLaboratory of Human Genetics of Infectious Diseases, Necker Branch, Paris Descartes University, Institut National de la Santé et de la Recherche Médicale U980, Necker Medical School, 75015 Paris, France; cPlateforme Bioinformatique, Université Paris Descartes, 75116 Paris, France; dDepartment of Computer Science, Ben-Gurion University of the Negev, Beer-Sheva 84105, Israel; eUnit of Human Evolutionary Genetics, Centre National de la Recherche Scientifique, Unité de Recherche Associée 3012, Institut Pasteur, F-75015 Paris, France; and fPediatric Immunology-Hematology Unit, Necker Hospital for Sick Children, 75015 Paris, France Edited* by Bruce Beutler, University of Texas Southwestern Medical Center, Dallas, TX, and approved February 15, 2013 (received for review October 19, 2012) High-throughput genomic data reveal thousands of gene variants to detect a single mutated gene, with the other polymorphic genes per patient, and it is often difficult to determine which of these being of less interest. This goes some way to explaining why, variants underlies disease in a given individual. However, at the despite the abundance of NGS data, the discovery of disease- population level, there may be some degree of phenotypic homo- causing alleles from such data remains somewhat limited. geneity, with alterations of specific physiological pathways under- We developed the human gene connectome (HGC) to over- come this problem. -

Current Status of Fish Vaccines in Japan
Fish and Shellfish Immunology 95 (2019) 236–247 Contents lists available at ScienceDirect Fish and Shellfish Immunology journal homepage: www.elsevier.com/locate/fsi Full length article Current status of fish vaccines in Japan T ∗ Yuta Matsuura, Sachiko Terashima, Tomokazu Takano, Tomomasa Matsuyama Research Center of Fish Diseases, National Research Institute of Aquaculture, Japan Fisheries Research and Education Agency, Minami.-Ise, Mie, Japan ARTICLE INFO ABSTRACT Keywords: Aquaculture is an important industry in Japan for the sustainable production of fish. It contributes to the di- Aquaculture versity of Japanese traditional food culture, which uses fish such as “sushi” and “sashimi”. In the recent Fish diseases aquaculture setting in Japan, infectious diseases have been an unavoidable problem and have caused serious Fish vaccines economic losses. Therefore, there is an urgent need to overcome the disease problem to increase the productivity Bacterial hemolytic jaundice of aquaculture. Although our country has developed various effective vaccines against fish pathogens, which Bacterial cold-water disease have contributed to disease prevention on fish farms, infectious diseases that cannot be controlled by conven- Erythrocytic inclusion body syndrome ff Nocardia tional inactivated vaccines are still a problem. Therefore, other approaches to developing e ective vaccines Piscine orthoreovirus other than inactivated vaccines are required. This review introduces the vaccine used in Japan within the context Plecoglossus altivelis poxvirus-like virus of the current status of finfish aquacultural production and disease problems. This review also summarizes the current research into vaccine development and discusses the future perspectives of fish vaccines, focusing on the problems associated with vaccine promotion in Japan. -

Miamiensis Avidus
Parasitology Research https://doi.org/10.1007/s00436-018-6010-8 ORIGINAL PAPER Development of a safe antiparasitic against scuticociliates (Miamiensis avidus) in olive flounders: new approach to reduce the toxicity of mebendazole by material remediation technology using full-overlapped gravitational field energy Jung-Soo Seo1 & Na-Young Kim2 & Eun-Ji Jeon2 & Nam-Sil Lee2 & En-Hye Lee3 & Myoung-Sug Kim2 & Hak-Je Kim4 & Sung-Hee Jung2 Received: 5 March 2018 /Accepted: 6 July 2018 # The Author(s) 2018 Abstract The olive flounder (Paralychthys olivaceus) is a representative farmed fish species in South Korea, which is cultured in land-based tanks and accounts for approximately 50% of total fish farming production. However, farmed olive flounder are susceptible to infection with parasitic scuticociliates, which cause scuticociliatosis, a disease resulting in severe economic losses. Thus, there has been a longstanding imperative to develop a highly stable and effective antiparasitic drug that can be rapidly administered, both orally and by bath, upon infection with scuticociliates. Although the efficacy of commercially available mebendazole (MBZ) has previously been established, this compound cannot be used for olive flounder due to hematological, biochemical, and histopathological side effects. Thus, we produced material remediated mebendazole (MR MBZ), in which elements comprising the molecule wereARTICLE remediated by using full-overlapped grav- itational field energy, thereby reducing the toxicity of the parent material. The antiparasitic effect of MR MBZ against scuticociliates in olive flounder was either similar to or higher than that of MBZ under the same conditions. Oral (100 and500mg/kgB.W.)andbath(100and500mg/L) administrations of MBZ significantly (p < 0.05) increased the values of hematological and biochemical parameters, whereas these values showed no increase in the MR MBZ administration group.