Trna-Isoleucine-Tryptophan Composite Gene
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Effect of Peptide Histidine Isoleucine on Water and Electrolyte Transport in the Human Jejunum
Gut: first published as 10.1136/gut.25.6.624 on 1 June 1984. Downloaded from Gut, 1984, 25, 624-628 Alimentary tract and pancreas Effect of peptide histidine isoleucine on water and electrolyte transport in the human jejunum K J MORIARTY, J E HEGARTY, K TATEMOTO, V MUTT, N D CHRISTOFIDES, S R BLOOM, AND J R WOOD From the Department of Gastroenterology, St Bartholomew's Hospital, London, The Liver Unit, King's College Hospital, London, Department ofMedicine, Hammersmith Hospital, London, and Department of Biochemistry, Karolinska Institute, Stockholm, Sweden SUMMARY Peptide histidine isoleucine, a 27 amino acid peptide with close amino acid sequence homology to vasoactive intestinal peptide and secretin, is distributed throughout the mammalian intestinal tract, where it has been localised to intramural neurones. An intestinal perfusion technique has been used to study the effect of intravenous peptide histidine isoleucine (44.5 pmol/kg/min) on water and electrolyte transport from a plasma like electrolyte solution in human jejunum in vivo. Peptide histidine isoleucine infusion produced peak plasma peptide histidine isoleucine concentrations in the range 2000-3000 pmolIl, flushing, tachycardia and a reduction in diastolic blood pressure. Peptide histidine isoleucine caused a significant inhibition of net absorption of water, sodium, potassium and bicarbonate and induced a net secretion of chloride, these changes being completely reversed during the post-peptide histidine isoleucine period. These findings suggest that endogenous peptide histidine isoleucine may participate in the neurohumoral regulation of water and electrolyte transport in the human jejunum. http://gut.bmj.com/ Peptide histidine isoleucine, isolated originally from INTESTINAL PERFUSION mammalian small intestine, is a 27-amino acid After an eight hour fast, each subject swallowed a peptide having close amino acid sequence homology double lumen intestinal perfusion tube, incorpo- to vasoactive intestinal peptide and secretin. -

Isoleucine, an Essential Amino Acid, Prevents Liver Metastases of Colon Cancer by Antiangiogenesis Kazumoto Murata1 and Masami Moriyama2
Research Article Isoleucine, an Essential Amino Acid, Prevents Liver Metastases of Colon Cancer by Antiangiogenesis Kazumoto Murata1 and Masami Moriyama2 1The First Department of Internal Medicine, Mie University School of Medicine, Tsu, Mie, Japan and 2Microbiology and Immunology, Keio University School of Medicine, Tokyo, Japan Abstract infection in the airways of cystic fibrosis (8), and susceptibility to salmonella infection in mouse intestinal tracts (9). In addition to In spite of recent advances in the treatment of colon cancer, h multiple liver metastases of colon cancer are still difficult to their direct antimicrobial activities, -defensins are strong treat. Some chemotherapeutic regimens have been reported to chemotactic factors for memory T cells and dendritic cells, be efficient, but there is a high risk of side effects associated suggesting that they also play an important role in acquired immunity (10–12). h-defensins are also inducible by inflammatory with these. Here, we show that isoleucine, an essential amino a h acid, prevents liver metastases in a mouse colon cancer cytokines, such as tumor necrosis factor- and interleukin-1 (13, 14). Recently, Fehlbaum et al. (15) reported that isoleucine metastatic model. Because isoleucine is a strong inducer of h B-defensin, we first hypothesized that it prevented liver and its analogues are highly specific inducers of -defensins. We thus originally hypothesized that isoleucine may contribute to metastases via the accumulation of dendritic cells or memory B tumor immunity through both innate and acquired immunity by T cells through up-regulation of -defensin. However, neither h B-defensin nor immunologic responses were induced by induction of -defensins. -
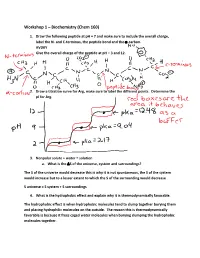
Workshop 1 – Biochemistry (Chem 160)
Workshop 1 – Biochemistry (Chem 160) 1. Draw the following peptide at pH = 7 and make sure to include the overall charge, label the N- and C-terminus, the peptide bond and the -carbon. AVDKY Give the overall charge of the peptide at pH = 3 and 12. 2. Draw a titration curve for Arg, make sure to label the different points. Determine the pI for Arg. 3. Nonpolar solute + water = solution a. What is the S of the universe, system and surroundings? The S of the universe would decrease this is why it is not spontaneous, the S of the system would increase but to a lesser extent to which the S of the surrounding would decrease S universe = S system + S surroundings 4. What is the hydrophobic effect and explain why it is thermodynamically favorable. The hydrophobic effect is when hydrophobic molecules tend to clump together burying them and placing hydrophilic molecules on the outside. The reason this is thermodynamically favorable is because it frees caged water molecules when burying clumping the hydrophobic molecules together. 5. Urea dissolves very readily in water, but the solution becomes very cold as the urea dissolves. How is this possible? Urea dissolves in water because when dissolving there is a net increase in entropy of the universe. The heat exchange, getting colder only reflects the enthalpy (H) component of the total energy change. The entropy change is high enough to offset the enthalpy component and to add up to an overall -G 6. A mutation that changes an alanine residue in the interior of a protein to valine is found to lead to a loss of activity. -

The Optimum Isoleucine:Lysine Ratio in Starter Diets to Maximize Growth Performance of the Early-Weaned Pig
Kansas Agricultural Experiment Station Research Reports Volume 0 Issue 10 Swine Day (1968-2014) Article 837 2000 The optimum isoleucine:lysine ratio in starter diets to maximize growth performance of the early-weaned pig B W. James Robert D. Goodband Michael D. Tokach See next page for additional authors Follow this and additional works at: https://newprairiepress.org/kaesrr Part of the Other Animal Sciences Commons Recommended Citation James, B W.; Goodband, Robert D.; Tokach, Michael D.; Nelssen, Jim L.; and DeRouchey, Joel M. (2000) "The optimum isoleucine:lysine ratio in starter diets to maximize growth performance of the early-weaned pig," Kansas Agricultural Experiment Station Research Reports: Vol. 0: Iss. 10. https://doi.org/10.4148/ 2378-5977.6677 This report is brought to you for free and open access by New Prairie Press. It has been accepted for inclusion in Kansas Agricultural Experiment Station Research Reports by an authorized administrator of New Prairie Press. Copyright 2000 Kansas State University Agricultural Experiment Station and Cooperative Extension Service. Contents of this publication may be freely reproduced for educational purposes. All other rights reserved. Brand names appearing in this publication are for product identification purposes only. No endorsement is intended, nor is criticism implied of similar products not mentioned. K-State Research and Extension is an equal opportunity provider and employer. The optimum isoleucine:lysine ratio in starter diets to maximize growth performance of the early-weaned pig Abstract A total of 360 weanling pigs (initially 12.3 lb BW and approximately 18 d of age) was used in a 14-d growth assay to determine the optimal isoleucine:lysine ratio to maximize growth performance. -

Amino Acid Chemistry
Handout 4 Amino Acid and Protein Chemistry ANSC 619 PHYSIOLOGICAL CHEMISTRY OF LIVESTOCK SPECIES Amino Acid Chemistry I. Chemistry of amino acids A. General amino acid structure + HN3- 1. All amino acids are carboxylic acids, i.e., they have a –COOH group at the #1 carbon. 2. All amino acids contain an amino group at the #2 carbon (may amino acids have a second amino group). 3. All amino acids are zwitterions – they contain both positive and negative charges at physiological pH. II. Essential and nonessential amino acids A. Nonessential amino acids: can make the carbon skeleton 1. From glycolysis. 2. From the TCA cycle. B. Nonessential if it can be made from an essential amino acid. 1. Amino acid "sparing". 2. May still be essential under some conditions. C. Essential amino acids 1. Branched chain amino acids (isoleucine, leucine and valine) 2. Lysine 3. Methionine 4. Phenyalanine 5. Threonine 6. Tryptophan 1 Handout 4 Amino Acid and Protein Chemistry D. Essential during rapid growth or for optimal health 1. Arginine 2. Histidine E. Nonessential amino acids 1. Alanine (from pyruvate) 2. Aspartate, asparagine (from oxaloacetate) 3. Cysteine (from serine and methionine) 4. Glutamate, glutamine (from α-ketoglutarate) 5. Glycine (from serine) 6. Proline (from glutamate) 7. Serine (from 3-phosphoglycerate) 8. Tyrosine (from phenylalanine) E. Nonessential and not required for protein synthesis 1. Hydroxyproline (made postranslationally from proline) 2. Hydroxylysine (made postranslationally from lysine) III. Acidic, basic, polar, and hydrophobic amino acids A. Acidic amino acids: amino acids that can donate a hydrogen ion (proton) and thereby decrease pH in an aqueous solution 1. -

Recent Progress in the Microbial Production of Pyruvic Acid
fermentation Review Recent Progress in the Microbial Production of Pyruvic Acid Neda Maleki 1 and Mark A. Eiteman 2,* 1 Department of Food Science, Engineering and Technology, University of Tehran, Karaj 31587-77871, Iran; [email protected] 2 School of Chemical, Materials and Biomedical Engineering, University of Georgia, Athens, GA 30602, USA * Correspondence: [email protected]; Tel.: +1-706-542-0833 Academic Editor: Gunnar Lidén Received: 10 January 2017; Accepted: 6 February 2017; Published: 13 February 2017 Abstract: Pyruvic acid (pyruvate) is a cellular metabolite found at the biochemical junction of glycolysis and the tricarboxylic acid cycle. Pyruvate is used in food, cosmetics, pharmaceutical and agricultural applications. Microbial production of pyruvate from either yeast or bacteria relies on restricting the natural catabolism of pyruvate, while also limiting the accumulation of the numerous potential by-products. In this review we describe research to improve pyruvate formation which has targeted both strain development and process development. Strain development requires an understanding of carbohydrate metabolism and the many competing enzymes which use pyruvate as a substrate, and it often combines classical mutation/isolation approaches with modern metabolic engineering strategies. Process development requires an understanding of operational modes and their differing effects on microbial growth and product formation. Keywords: auxotrophy; Candida glabrata; Escherichia coli; fed-batch; metabolic engineering; pyruvate; pyruvate dehydrogenase 1. Introduction Pyruvic acid (pyruvate at neutral pH) is a three carbon oxo-monocarboxylic acid, also known as 2-oxopropanoic acid, 2-ketopropionic acid or acetylformic acid. Pyruvate is biochemically located at the end of glycolysis and entry into the tricarboxylic acid (TCA) cycle (Figure1). -

Radhakrishnan, Iii 4
NATURE OF THE GENETIC BLOCKS IN THE ISOLEUCINE-VALINE MUTANTS OF SALMONELLA R. P. WAGNER AND ARLOA BERGQUIST The Genetics Laboratory of the Department of Zoology, The Uniuersity of Texas, Austin, Texas Received April 26, 1960 HE metabolic pathways leading to the biosynthesis of isoleucine and valine are now sufficiently well understood, as a result of the work of a number of investigators ( STRASSMAN,THOMAS and WEINHOUSE1955; STRASSMAN,THOMAS, LOCKEand WEINHOUSE1956; STRASSMAN,SHATTON, CORSEY and WEINHOUSE 1958; UMBARGER1958a,b; ADELBERG1955; WAGNER,RADHAKRISHNAN and SNELL1958; RADHAKRISHNAN,WAGNER and SNELL1960; and RADHAKRISHNAN and SNELL1960) to make it possible to investigate the nature of the genetic blocks in mutant organisms requiring isoleucine and valine. This communication describes the results of the investigation of a series of mutants of Salmonella typhimurium originally isolated in the laboratory of DR. M. DEMEREC,The Carnegie Institute of Washington, Cold Spring Harbor, New York. It is limited to the purely biochemical aspects of these mutants, but is ,preceded by a com- munication from GLANVILLEand DEMEREC1960, which describes the linkage studies made with these mutants, and correlates the genetic with the biochemical 3ata. The biosynthesis of isoleucine and valine is believed to occur as shown in Figure 1. In the work to be described here only the steps proceeding from the 3-keto acids, a-acetolactic acid and a-aceto-P-hydroxybutyricacid, have been :onsidered in detail. The following abbreviations are used in Figure I and in mbsequent parts of this communication: AHB = a-aceto-a-hydroxybutyric acid; CHa C Ha CHa CHa I 1 I I c*o CHa-C-OH CHfC-OH CHI-C-H 1 I TPNH 1 I c.0 ~ *Valine I COOH COOH I COOH I COOH (PYRUVIC ACID) (ALI I (HKVI I (DHV) I I I I I I I I + Pyruvic Acid Step1 StepII Stepm Step H 4 I I I I cn3 CH, I CH3 I I I I I CHZ C=O I CH~CH~C-OH Threonine-GO I ~CH3CH2-~-OH-C=0I+! I I I I COOH COOH COOH COOH COOH (U-kmtobutyric acid1 (AH01 (HKII (DUI) (KI) FIGURE1 .-The biosynthetic pathway leading to isoleucine and valine. -

Determination of Branched-Chain Keto Acids in Serum and Muscles Using High Performance Liquid Chromatography-Quadrupole Time-Of-Flight Mass Spectrometry
molecules Article Determination of Branched-Chain Keto Acids in Serum and Muscles Using High Performance Liquid Chromatography-Quadrupole Time-of-Flight Mass Spectrometry You Zhang, Bingjie Yin, Runxian Li and Pingli He * State Key Laboratory of Animal Nutrition, College of Animal Science and Technology, China Agricultural University, Beijing 100193, China; [email protected] (Y.Z.); [email protected] (B.Y.); [email protected] (R.L.) * Correspondence: [email protected]; Tel.: +86-10-6273-3688 Received: 4 December 2017; Accepted: 8 January 2018; Published: 11 January 2018 Abstract: Branched-chain keto acids (BCKAs) are derivatives from the first step in the metabolism of branched-chain amino acids (BCAAs) and can provide important information on animal health and disease. Here, a simple, reliable and effective method was developed for the determination of three BCKAs (α-ketoisocaproate, α-keto-β-methylvalerate and α-ketoisovalerate) in serum and muscle samples using high performance liquid chromatography-quadrupole time-of-flight mass spectrometry (HPLC-Q-TOF/MS). The samples were extracted using methanol and separated on a 1.8 µm Eclipse Plus C18 column within 10 min. The mobile phase was 10 mmol L−1 ammonium acetate aqueous solution and acetonitrile. The results showed that recoveries for the three BCKAs ranged from 78.4% to 114.3% with relative standard deviation (RSD) less than 9.7%. The limit of quantitation (LOQ) were 0.06~0.23 µmol L−1 and 0.09~0.27 nmol g−1 for serum and muscle samples, respectively. The proposed method can be applied to the determination of three BCKAs in animal serum and muscle samples. -

The Isoleucine-Valine Biosynthesis Pathway in Staphylococcus Aureus David Lee Bronson Iowa State University
Iowa State University Capstones, Theses and Retrospective Theses and Dissertations Dissertations 1969 The isoleucine-valine biosynthesis pathway in Staphylococcus aureus David Lee Bronson Iowa State University Follow this and additional works at: https://lib.dr.iastate.edu/rtd Part of the Microbiology Commons Recommended Citation Bronson, David Lee, "The isoleucine-valine biosynthesis pathway in Staphylococcus aureus " (1969). Retrospective Theses and Dissertations. 3631. https://lib.dr.iastate.edu/rtd/3631 This Dissertation is brought to you for free and open access by the Iowa State University Capstones, Theses and Dissertations at Iowa State University Digital Repository. It has been accepted for inclusion in Retrospective Theses and Dissertations by an authorized administrator of Iowa State University Digital Repository. For more information, please contact [email protected]. This dissertation has been microfilmed exactly as received 70-7679 BRONSON, David Lee, 1936- THE ISOLEUCINE-VALINE BIOSYNTHESIS PATHWAY IN STAPHYLOCOCCUS AUREUS. Iowa State University, Ph.D., 1969 Bacteriology University Microfilms, Inc., Ann Arbor, Michigan THE ISOLEUCINE-VALINE BIOSYNTHESIS PATHWAY IN STAPHYLOCOCCUS AUREUS by David Lee Bronson A Dissertation Submitted to the Graduate Faculty in Partial Fulfillment of The Requirements for the Degree of DOCTOR OF PHILOSOPHY Major Subject: Bacteriology Approved: Signature was redacted for privacy. In Ch of Major Work Signature was redacted for privacy. Signature was redacted for privacy. Iowa State University -

Omega-3 Fatty Acids Correlate with Gut Microbiome Diversity And
Omega-3 fatty acids correlate with gut microbiome diversity and production of N- carbamylglutamate in middle aged and elderly women Cristina Mennia, Jonas Zierera, Tess Pallistera, Matthew A Jacksona, Tao Longb, Robert P Mohneyc, Claire J Stevesa, Tim D Spectora, Ana M Valdesa,d,e a Department of Twin Research and Genetic Epidemiology, Kings College London, London, UK. b Sanford Burnham Prebys, USA cMetabolon Inc., Raleigh-Durham, NC 27709, USA. dSchool of Medicine , Nottingham City Hospital, Hucknall Road, Nottingham, UK. e NIHR Nottingham Biomedical Research Centre, Nottingham, UK. Corresponding author: Dr Ana M Valdes School of Medicine Clinical Sciences Building, Nottingham City Hospital, Hucknall Road, Nottingham, NG5 1PB, UK Phone number: +44 (0)115 823 1954; Fax number:+44(0) 115 823 1757 email: [email protected] Supplementary Table 1. List of faecal metabolites significantly associated to DHA circulating levels (FDR<0.05)adjusting for age, BMI and family relatedness. Super Pathway Sub pathway Metabolite BETA SE P Q Lipid Polyunsaturated Fatty Acid (n3 and n6) eicosapentaenoate (EPA; 20:5n3) 0.15 0.04 4.35x10-5 0.01 Xenobiotics Carbamylated aminoacid N-carbamylglutamate 0.15 0.04 1.21x10-4 0.02 Peptide Dipeptide Derivative anserine 0.13 0.04 5.39x10-4 0.04 Supplementary Table 2. Associations between five measures of microbiome diversity and circulating levels of PUFA adjusting for age, BMI fibre intake and family relatedness SHANNON DIVERSITY CHAO1 OBSERVED Phylogenetic Diversity SIMPSON SPECIES Beta SE P Beta SE P Beta SE P Beta SE P Beta SE P DHA 0.13 0.04 0.002 0.12 0.04 0.003 0.14 0.04 0.001 0.12 0.04 0.01 0.09 0.04 0.02 FAW3 0.12 0.04 0.005 0.13 0.04 0.003 0.13 0.04 0.002 0.12 0.05 0.01 0.08 0.04 0.05 LA 0.10 0.05 0.02 0.10 0.05 0.03 0.10 0.05 0.03 0.10 0.05 0.03 0.09 0.04 0.04 (18:2) FAW6 0.09 0.04 0.02 0.10 0.04 0.01 0.10 0.04 0.01 0.10 0.04 0.02 0.08 0.04 0.05 DHA=22:6 docosahexaenoic acid;, FAW3= Omega-3 fatty acids; 18:2, LA= linoleic acid;; FAW6=omega-6 fatty acids Supplementary Table 3. -

Safety Assessment of Α-Amino Acids As Used in Cosmetics
Safety Assessment of α-Amino Acids as Used in Cosmetics Status: Final Report for Public Distribution Release Date: October 5, 2012 Panel Meeting Date: September 10-11, 2012 The 2012 Cosmetic Ingredient Review Expert Panel members are: Chairman, Wilma F. Bergfeld, M.D., F.A.C.P.; Donald V. Belsito, M.D.; Curtis D. Klaassen, Ph.D.; Daniel C. Liebler, Ph.D.; Ronald A. Hill, Ph.D. James G. Marks, Jr., M.D.; Ronald C. Shank, Ph.D.; Thomas J. Slaga, Ph.D.; and Paul W. Snyder, D.V.M., Ph.D. The CIR Director is F. Alan Andersen, Ph.D. This safety assessment was prepared by Christina L. Burnett, Scientific Analyst/Writer, and Bart Heldreth, Ph.D., Chemist CIR. © Cosmetic Ingredient Review 1101 17th Street, NW, Suite 412 Washington, DC 20036-4702 ph 202.331.0651 fax 202.331.0088 [email protected] 1 ABSTRACT The Cosmetic Ingredient Review Expert Panel (the Panel) reviewed the safety of α-amino acids, which function primarily as hair and skin conditioning agents in cosmetic products. The safety of α-amino acids as direct food additives has been well established based on extensive research through acute and chronic dietary exposures. The Panel focused its review on dermal irritation and sensitization data relevant to the use of these ingredients in topical cosmetics. The Panel concluded that α-amino acids were safe as cosmetic ingredients in the practices of use and concentration of this safety assessment. INTRODUCTION Amino acids and their salts are widely used as cosmetic ingredients, and function primarily as hair conditioning agents and skin conditioning agents (humectant and miscellaneous). -

Utilization of Alpha-Ketoisocaproate for Protein Synthesis in Uremic Rats
CORE Metadata, citation and similar papers at core.ac.uk Provided by Elsevier - Publisher Connector Kidney International, Vol. 30 (1986), pp. 891—894 Utilization of alpha-ketoisocaproate for protein synthesis in uremic rats KRIANG TUNGSANGA, CHANG W. KANG, and MACKENZIE WALSER Department of Pharmacology and Molecular Science, and Department of Medicine, Johns Hopkins University School of Medicine, Baltimore, Maryland U.S.A. Utilization of alpha-.ketoisocaproate for protein synthesis in uremic logues [1]. The reason for this was that studies of isolated rats. We have recently shown that the nutritional efficiency, R, ofperfused rat liver had shown that substantial fractions of each alpha—ketoisocaproate (KIC) as a substitute for leucine, defined as the ratio of the dose of leucine to the dose of KIC (on a leucine—free diet) BCKA were oxidized rather than transaminated [8]. for equal growth, can be evaluated isotopically: '4C-KIC and 3H-leucine Chawla, Stackhouse and Wadsworth [9] compared the are administered p.o.; six hours later, '4C/3H in the leucine of whole growth of rats fed aipha—ketoisocaproate (KIC) in place of body protein, divided by '4CPH in the injectate, gives a value indistin- leucine, and reported that four to five times as much KIC was guishable from R assessed in the same animals by growth experiments. To see how chronic uremia affects R, 11/12 nephrectomized rats and required to achieve equal growth. They defined nutritional sham—operated controls were fed a regular diet for 15 days and then efficiency of KIC as the ratio of the dose of leucine to the dose given these isotopes p.o.