Implicit Modeling of the Impact of Adsorption on Solid Surfaces For
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

CYP2A6) by P53
Transcriptional Regulation of Human Stress Responsive Cytochrome P450 2A6 (CYP2A6) by p53 Hao Hu M.Biotech. (Biotechnology) 2012 The University of Queensland B.B.A. 2009 University of Electronic Science and Technology of China B.Sc. (Pharmacy) 2009 University of Electronic Science and Technology of China A thesis submitted for the degree of Doctor of Philosophy at The University of Queensland in 2016 School of Medicine ABSTRACT Human cytochrome P450 (CYP) 2A6 is highly expressed in the liver and the encoding gene is regulated by various stress activated transcription factors, such as the nuclear factor (erythroid-derived 2)-like 2 (Nrf-2). Unlike the other xenobiotic metabolising CYP enzymes (XMEs), CYP2A6 only plays a minor role in xenobiotic metabolism. The CYP2A6 is highly induced by multiple forms of cellular stress conditions, where XMEs expression is normally inhibited. Recent findings suggest that the CYP2A6 plays an important role in regulating BR homeostasis. A computer based sequence analysis on the 3 kb proximate CYP2A6 promoter revealed several putative binding sites for p53, a protein that mediates regulation of antioxidant and apoptosis pathways. In this study, the role of p53 in CYP2A6 gene regulation is demonstrated. The site closest to transcription start site (TSS) is highly homologous with the p53 consensus sequence. The p53 responsiveness of this site was confirmed by transfections with various stepwise deleted of CYP2A6-5’-Luc constructs containing the putative p53RE. Deletion of the putative p53RE resulted in a total abolishment of p53 responsiveness of CYP2A6 promoter. Specific binding of p53 to the putative p53RE was detected by electrophoresis mobility shift assay. -

Characterisation of Bilirubin Metabolic Pathway in Hepatic Mitochondria Siti Nur Fadzilah Muhsain M.Sc
Characterisation of Bilirubin Metabolic Pathway in Hepatic Mitochondria Siti Nur Fadzilah Muhsain M.Sc. (Medical Research) 2005 Universiti Sains Malaysia Postgrad. Dip. (Toxicology) 2003 University of Surrey B.Sc.(Biomed. Sc.) 2000 Universiti Putra Malaysia A thesis submitted for the degree of Doctor of Philosophy at The University of Queensland in 2014 School of Medicine ABSTRACT Bilirubin (BR), a toxic waste product of degraded haem, is a potent antioxidant at physiological concentrations. To achieve the maximum benefit of BR, its intracellular level needs to be carefully regulated. A system comprising of two enzymes, haem oxygenase-1 (HMOX1) and cytochrome P450 2A5 (CYP2A5) exists in the endoplasmic reticulum (ER), responsible for regulating BR homeostasis. This system is induced in response to oxidative stress. In this thesis, oxidative stress caused accumulation of these enzymes in mitochondria — major producers and targets of reactive oxygen species (ROS) — is demonstrated. To understand the significance of this intracellular targeting, properties of microsomal and mitochondrial BR metabolising enzymes were compared and the capacity of mitochondrial CYP2A5 to oxidise BR in response to oxidative stress is reported. Microsomes and mitochondrial fractions were isolated from liver homogenates of DBA/2J mice, administered with sub-toxic dose of pyrazole, an oxidant stressor. The purity of extracted organelles was determined by analysing the expressions and activities of their respective marker enzymes. HMOX1 and CYP2A5 were significantly increased in microsomes and even more so in mitochondria in response to pyrazole-induced oxidative stress. By contrast, the treatment did not increase either microsomes or mitochondrial Uridine-diphosphate-glucuronosyltransferase 1A1 (UGT1A1), the sole enzyme that catalyses BR elimination through glucuronidation. -

Dissertation
Dissertation Submitted to the Combined Faculties for the Natural Sciences and for Mathematics of the Ruperto-Carola University of Heidelberg, Germany for the Degree of Doctor of Natural Sciences Presented by Ann-Cathrin Hofer (M.Sc.) Born in Heidelberg, Germany Oral Examination: 12th of September 2016 Regulatory T cells protect the neonatal liver and secure the hepatic circadian rhythm Referees 1st Referee: Prof. Dr. Peter Angel 2nd Referee: Dr. Markus Feuerer This dissertation was performed and written during the period from November 2012 to May 2016 in the German Cancer Research Center (DKFZ) under the supervision of Prof. Dr. Peter Angel and direct supervision of Dr. Markus Feuerer. The dissertation was submitted to the Combined Faculties for the Natural Sciences and for Mathematics of the Ruperto-Carola University of Heidelberg, Germany in June 2016. German Cancer Research Center (DKFZ) Immune Tolerance (D100) Im Neuenheimer Feld 280 69120 Heidelberg, Germany I II Confirmation Hereby, I confirm that I have written this thesis independently, using only the results of my investigation unless otherwise stated. Furthermore, I declare that I have not submitted this thesis for a degree to any other academic or similar institution. Parts of this dissertation have been submitted for publishing: Regulatory T cells protect the liver early in life and safeguard the hepatic circadian rhythm Ann-Cathrin Hofer, Thomas Hielscher, David M. Richards, Michael Delacher, Ulrike Träger, Sophia Föhr, Artyom Vlasov, Marvin Wäsch, Marieke Esser, Annette Kopp-Schneider, Achim Breiling, Frank Lyko, Ursula Klingmüller, Peter Angel, Jakub Abramson, Jeroen Krijgsveld & Markus Feuerer Parts of the experiments in this dissertation were performed in collaboration with other research groups as follows: CG methylation analysis with the 454 pyrosequencing technology: Division of Epigenetics, DKFZ, Heidelberg Dr. -

Solarbio Catalogue with PRICES
CAS Name Grade Purity Biochemical Reagent Biochemical Reagent 75621-03-3 C8390-1 3-((3-Cholamidopropyl)dimethylammonium)-1-propanesulfonateCHAPS Ultra Pure Grade 1g 75621-03-3 C8390-5 3-((3-Cholamidopropyl)dimethylammonium)-1-propanesulfonateCHAPS 5g 57-09-0 C8440-25 Cetyl-trimethyl Ammonium Bromide CTAB High Pure Grade ≥99.0% 25g 57-09-0 C8440-100 Cetyl-trimethyl Ammonium Bromide CTAB High Pure Grade ≥99.0% 100g 57-09-0 C8440-500 Cetyl-trimethyl Ammonium Bromide CTAB High Pure Grade ≥99.0% 500g E1170-100 0.5M EDTA (PH8.0) 100ml E1170-500 0.5M EDTA (PH8.0) 500ml 6381-92-6 E8030-100 EDTA disodium salt dihydrate EDTA Na2 Biotechnology Grade ≥99.0% 100g 6381-92-6 E8030-500 EDTA disodium salt dihydrate EDTA Na2 Biotechnology Grade ≥99.0% 500g 6381-92-6 E8030-1000 EDTA disodium salt dihydrate EDTA Na2 Biotechnology Grade ≥99.0% 1kg 6381-92-6 E8030-5000 EDTA disodium salt dihydrate EDTA Na2 Biotechnology Grade ≥99.0% 5kg 60-00-4 E8040-100 Ethylenediaminetetraacetic acid EDTA Ultra Pure Grade ≥99.5% 100g 60-00-4 E8040-500 Ethylenediaminetetraacetic acid EDTA Ultra Pure Grade ≥99.5% 500g 60-00-4 E8040-1000 Ethylenediaminetetraacetic acid EDTA Ultra Pure Grade ≥99.5% 1kg 67-42-5 E8050-5 Ethylene glycol-bis(2-aminoethylether)-N,N,NEGTA′,N′-tetraacetic acid Ultra Pure Grade ≥97.0% 5g 67-42-5 E8050-10 Ethylene glycol-bis(2-aminoethylether)-N,N,NEGTA′,N′-tetraacetic acid Ultra Pure Grade ≥97.0% 10g 50-01-1 G8070-100 Guanidine Hydrochloride Guanidine HCl ≥98.0%(AT) 100g 50-01-1 G8070-500 Guanidine Hydrochloride Guanidine HCl ≥98.0%(AT) 500g 56-81-5 -

1 Molecular Physiology and Pathophysiology of Bilirubin Handling by the Blood, Liver
1 1 MOLECULAR PHYSIOLOGY AND PATHOPHYSIOLOGY OF BILIRUBIN HANDLING BY THE BLOOD, LIVER, 2 INTESTINE, AND BRAIN IN THE NEWBORN 3 THOR W.R. HANSEN1, RONALD J. WONG2, DAVID K. STEVENSON2 4 1Division of Paediatric and Adolescent Medicine, Institute of Clinical Medicine, Faculty of Medicine, 5 University of Oslo, Norway 6 2Department of Pediatrics, Stanford University School of Medicine, Stanford CA, USA 7 __________________________________________________________________________________ 8 2 9 I. Introduction 10 II. Bilirubin in the Body 11 A. Bilirubin Chemistry 12 1. Bilirubin structure 13 2. Bilirubin solubility 14 3. Bilirubin isomers 15 4. Heme degradation 16 5. Biliverdin and biliverdin reductase (BVR) 17 B. Bilirubin as an Antioxidant 18 C. Bilirubin as a Toxin 19 1. Bilirubin effects on enzyme activity 20 2. Toxicity of bilirubin conjugates and isomers 21 D. Other Functions/Roles 22 1. Drug displacement by bilirubin 23 2. Bilirubin interactions with the immune system and 24 inflammatory/infectious mechanisms 25 III. The Production of Bilirubin in the Body 26 A. Heme Catabolism and Its Regulation 27 1. Genetic variants in bilirubin production 28 B. The Effect of Hemolysis 29 1. Disorders associated with increased bilirubin production 30 IV. Bilirubin Binding and Transport in Blood 31 V. Bilirubin in the Liver 32 A. Hepatocellular Uptake and Intracellular Processing 33 B. Bilirubin Conjugation 34 1. Genetic variants in bilirubin conjugation 3 35 a. Crigler-Najjar syndrome type I 36 b. Crigler-Najjar syndrome type II 37 c. Gilbert syndrome 38 2. Genetic variants in transporter proteins 39 C. Bilirubin Excretion 40 VI. Bilirubin in the Intestines 41 A. -

Junbai Li Editor Supramolecular Chemistry of Biomimetic Systems Supramolecular Chemistry of Biomimetic Systems Junbai Li Editor
Junbai Li Editor Supramolecular Chemistry of Biomimetic Systems Supramolecular Chemistry of Biomimetic Systems Junbai Li Editor Supramolecular Chemistry of Biomimetic Systems 123 Editor Junbai Li Institute of Chemistry, Chinese Academy of Sciences Beijing China ISBN 978-981-10-6058-8 ISBN 978-981-10-6059-5 (eBook) DOI 10.1007/978-981-10-6059-5 Library of Congress Control Number: 2017949116 © Springer Nature Singapore Pte Ltd. 2017 This work is subject to copyright. All rights are reserved by the Publisher, whether the whole or part of the material is concerned, specifically the rights of translation, reprinting, reuse of illustrations, recitation, broadcasting, reproduction on microfilms or in any other physical way, and transmission or information storage and retrieval, electronic adaptation, computer software, or by similar or dissimilar methodology now known or hereafter developed. The use of general descriptive names, registered names, trademarks, service marks, etc. in this publication does not imply, even in the absence of a specific statement, that such names are exempt from the relevant protective laws and regulations and therefore free for general use. The publisher, the authors and the editors are safe to assume that the advice and information in this book are believed to be true and accurate at the date of publication. Neither the publisher nor the authors or the editors give a warranty, express or implied, with respect to the material contained herein or for any errors or omissions that may have been made. The publisher remains neutral with regard to jurisdictional claims in published maps and institutional affiliations. Printed on acid-free paper This Springer imprint is published by Springer Nature The registered company is Springer Nature Singapore Pte Ltd. -
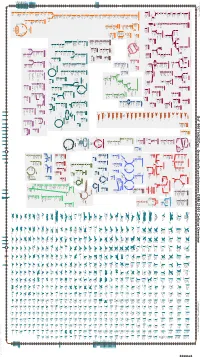
Generated by SRI International Pathway Tools Version 25.0 on Sat
Authors: Pallavi Subhraveti Ron Caspi Quang Ong Peter D Karp An online version of this diagram is available at BioCyc.org. Biosynthetic pathways are positioned in the left of the cytoplasm, degradative pathways on the right, and reactions not assigned to any pathway are in the far right of the cytoplasm. Transporters and membrane proteins are shown on the membrane. Ingrid Keseler Periplasmic (where appropriate) and extracellular reactions and proteins may also be shown. Pathways are colored according to their cellular function. Gcf_900114625Cyc: Granulicella pectinivorans DSM 21001 Cellular Overview Connections between pathways are omitted for legibility. -

Supplemental Table S1: Comparison of the Deleted Genes in the Genome-Reduced Strains
Supplemental Table S1: Comparison of the deleted genes in the genome-reduced strains Legend 1 Locus tag according to the reference genome sequence of B. subtilis 168 (NC_000964) Genes highlighted in blue have been deleted from the respective strains Genes highlighted in green have been inserted into the indicated strain, they are present in all following strains Regions highlighted in red could not be deleted as a unit Regions highlighted in orange were not deleted in the genome-reduced strains since their deletion resulted in severe growth defects Gene BSU_number 1 Function ∆6 IIG-Bs27-47-24 PG10 PS38 dnaA BSU00010 replication initiation protein dnaN BSU00020 DNA polymerase III (beta subunit), beta clamp yaaA BSU00030 unknown recF BSU00040 repair, recombination remB BSU00050 involved in the activation of biofilm matrix biosynthetic operons gyrB BSU00060 DNA-Gyrase (subunit B) gyrA BSU00070 DNA-Gyrase (subunit A) rrnO-16S- trnO-Ala- trnO-Ile- rrnO-23S- rrnO-5S yaaC BSU00080 unknown guaB BSU00090 IMP dehydrogenase dacA BSU00100 penicillin-binding protein 5*, D-alanyl-D-alanine carboxypeptidase pdxS BSU00110 pyridoxal-5'-phosphate synthase (synthase domain) pdxT BSU00120 pyridoxal-5'-phosphate synthase (glutaminase domain) serS BSU00130 seryl-tRNA-synthetase trnSL-Ser1 dck BSU00140 deoxyadenosin/deoxycytidine kinase dgk BSU00150 deoxyguanosine kinase yaaH BSU00160 general stress protein, survival of ethanol stress, SafA-dependent spore coat yaaI BSU00170 general stress protein, similar to isochorismatase yaaJ BSU00180 tRNA specific adenosine -

From Heterogeneous and Extracellular Electron Transfer to Applications in Aqueous Batteries and Biofuel Cells
UNIVERSIDADE DE SÃO PAULO INSTITUTO DE QUÍMICA DE SÃO CARLOS PROGRAMA DE PÓS-GRADUAÇÃO EM QUÍMICA Graziela Cristina Sedenho Bioelectrochemical and bioinspired energy conversion systems: from heterogeneous and extracellular electron transfer to applications in aqueous batteries and biofuel cells São Carlos – SP 2021 GRAZIELA CRISTINA SEDENHO Bioelectrochemical and bioinspired energy conversion systems: from heterogeneous and extracellular electron transfer to applications in aqueous batteries and biofuel cells Thesis presented to the São Carlos Institute of Chemistry (IQSC), University of São Paulo, in partial fulfillment of the requirements for the degree of Doctor of Science. Concentration area: Physical Chemistry Advisor: Prof. Dr. Frank Nelson Crespilho Exemplar revisado O exemplar original encontra-se em acervo reservado na Biblioteca do IQSC-USP São Carlos – SP 2021 Autorizo a reprodução e divulgação total ou parcial deste trabalho, por qualquer meio convencional ou eletrônico para fins de estudo e pesquisa, desde que citada a fonte. Assinatura: Data: 06/01/2021 Ficha Catalográfica elaborada pela Seção de Referência e Atendimento ao Usuário do SBI/IQSC Sedenho, Graziela Cristina Bioelectrochemical and bioinspired energy conversion systems: from heterogeneous and extracellular electron transfer to applications in aqueous batteries and biofuel cells / Graziela Cristina Sedenho. — São Carlos, 2021. 137 f. Tese (Doutorado em Físico-Química) — Instituto de Química de São Carlos / Universidade de São Paulo, 2021. Orientador: Prof. Dr. Frank Nelson Crespilho 1. Transformação de energia. 2. Eletrodos de carbono. 3. Imobilização enzimática. 4. Reação de redução de oxigênio. 5. Transferência extracelular de elétrons. I. Título. Wilneide do Carmo Marchi Maiorano - CRB: 3978/8 ACKNOWLEDGMENTS • To my parents, Luís and Sueli, for teaching me to be strong, for always supporting and encouraging me. -

Prediction of Anti-Hiv Activity of Flavanoid Constituents Through PASS
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by OpenSIUC Ethnobotanical Leaflets 12: 954-94. 2008. Prediction of Anti-Hiv Activity of Flavanoid Constituents Through PASS M. Maridass1, G. Raju2, K. Thangavel3 and S. Ghanthikumar4 1Animal Health Research Unit, St.Xavier’s College (Autonomous), Palayamkottai-627002 2Dept. of Advanced Zoology and Biotechnology, PioneerKumaraswamy College, Nagercoil, 629 002 3Dept. of Biotechnology, Sri Paramakalyani College, Alwarkurichi, Thirunelveli – 627 412 4Centre for Biodiversity and Biotechnology,St. Xavier’s College (Autonomous), Palayamkottai-02, Tamil Nadu, India E-mail: [email protected] Issued 08 November 2008 ABSTRACT Prediction of Activity Spectra for Substances (PASS), a new computer program, has been generated to evaluate the biological activity of four major flavanoids from a medicinal herb, Boesenbergia pandurata Holtt. (Zingiberaceae). Principal anti-HIV and other biological activities of pinostrobin, pinocembrin, cardamonin and alpinetin were predicted through PASS, their similarity and difference in the mechanisms of action with reference to accessory biological activities have been compared (Tables 1-4) and verified with the available data (http://195.178.207.233/PASS/socket1.php.) on pharmacological and toxicological activity of these compounds. KEY WORDS: Anti-HIV Drugs, Flavanoids, Boesenbergia pandurata, Pinostrobin, Pinocembrin, Cardamonin, Alpinetin. INTRODUCTION Acquired immunodeficiency syndrome (AIDS) has evolved rapidly into an epidemic and world-wide health crisis. The number of people infected with HIV rose to just over 40 million in 2006, an increase of over 2 million since 2004 (WHO/UNAIDS (2006). More than 60% of infected people live in sub-Saharan Africa, where at least 2 million deaths from HIV/AIDS occurred in 2006. -

Biochemical Characterization of Multicopper Oxidase Enzymes from Lignin Degrading Bacteria
A Thesis Submitted for the Degree of PhD at the University of Warwick Permanent WRAP URL: http://wrap.warwick.ac.uk/116117 Copyright and reuse: This thesis is made available online and is protected by original copyright. Please scroll down to view the document itself. Please refer to the repository record for this item for information to help you to cite it. Our policy information is available from the repository home page. For more information, please contact the WRAP Team at: [email protected] warwick.ac.uk/lib-publications Biochemical characterization of multicopper oxidase enzymes from lignin degrading bacteria Rommel Santiago Granja Trávez A thesis submitted in partial fulfilment of the requirements for the degree of Doctor of Philosophy in Chemistry University of Warwick Department of Chemistry October 2018 Supervisor: Professor Timothy D.H. Bugg I. Table of Contents I. Table of Contents ________________________________________________ i II. List of Figures ____________________________________________________ v III. List of Tables ________________________________________________ xiii IV. List of Abbreviations ___________________________________________ xv V. Acknowledgements ____________________________________________ xviii VI. Declaration _________________________________________________ xix VII. Scholarly outcomes ____________________________________________ xx VIII. Abstract ____________________________________________________ xxi 1 Introduction _____________________________________________________ 1 1.1 Lignin: function and -

Biochemical and Structural Characterisation of the Copper Containing
VTT SCIENCE 16 Biochemical and structural characterisation of the IENCE C • copper containing oxidoreductases catechol •S T S E N C oxidase, tyrosinase, and laccase from ascomycete O H I N S O fungi I V Dissertation L • O S G T 16 Catechol oxidase (EC 1.10.3.1), tyrosinase (EC 1.14.18.1), and laccase Y H • R G (EC 1.10.3.2) are copper-containing metalloenzymes. They oxidise I E L S H E G A I substituted phenols and use molecular oxygen as a terminal electron R H C H acceptor. Catechol oxidases and tyrosinases catalyse the oxidation of Biochemical and structural characterisation of the copper containing... p-substituted o-diphenols to the corresponding o-quinones. Tyrosinases also catalyse the introduction of a hydroxyl group in the ortho position of p-substituted monophenols and the subsequent oxidation to the corresponding o-quinones. Laccases can oxidise a wide range of compounds by removing single electrons from the reducing group of the substrate and generate free radicals. The reaction products of these oxidases can react further non-enzymatically and lead to formation of polymers and cross-linking of proteins and carbohydrates, in certain conditions. The work focused on examination of the properties of catechol oxidases, tyrosinases and laccases. A novel catechol oxidase from the ascomycete fungus Aspergillus oryzae was characterised from biochemical and structural point of view. Tyrosinases from Trichoderma reesei and Agaricus bisporus were examined in terms of substrate specificity and inhibition. The oxidation capacity of laccases was elucidated by using a set of laccases with different redox potential and a set of substituted phenolic substrates with different redox potential.