Lab-Made Electronic Nose for Fast Detection of Listeria Monocytogenes and Bacillus Cereus
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Food Poisoning Toxins of Bacillus Cereus
toxins Review The Food Poisoning Toxins of Bacillus cereus Richard Dietrich 1,†, Nadja Jessberger 1,*,†, Monika Ehling-Schulz 2 , Erwin Märtlbauer 1 and Per Einar Granum 3 1 Department of Veterinary Sciences, Faculty of Veterinary Medicine, Ludwig Maximilian University of Munich, Schönleutnerstr. 8, 85764 Oberschleißheim, Germany; [email protected] (R.D.); [email protected] (E.M.) 2 Department of Pathobiology, Functional Microbiology, Institute of Microbiology, University of Veterinary Medicine Vienna, 1210 Vienna, Austria; [email protected] 3 Department of Food Safety and Infection Biology, Faculty of Veterinary Medicine, Norwegian University of Life Sciences, P.O. Box 5003 NMBU, 1432 Ås, Norway; [email protected] * Correspondence: [email protected] † These authors have contributed equally to this work. Abstract: Bacillus cereus is a ubiquitous soil bacterium responsible for two types of food-associated gastrointestinal diseases. While the emetic type, a food intoxication, manifests in nausea and vomiting, food infections with enteropathogenic strains cause diarrhea and abdominal pain. Causative toxins are the cyclic dodecadepsipeptide cereulide, and the proteinaceous enterotoxins hemolysin BL (Hbl), nonhemolytic enterotoxin (Nhe) and cytotoxin K (CytK), respectively. This review covers the current knowledge on distribution and genetic organization of the toxin genes, as well as mechanisms of enterotoxin gene regulation and toxin secretion. In this context, the exceptionally high variability of toxin production between single strains is highlighted. In addition, the mode of action of the pore-forming enterotoxins and their effect on target cells is described in detail. The main focus of this review are the two tripartite enterotoxin complexes Hbl and Nhe, but the latest findings on cereulide and CytK are also presented, as well as methods for toxin detection, and the contribution of further putative virulence factors to the diarrheal disease. -

The Influence of Probiotics on the Firmicutes/Bacteroidetes Ratio In
microorganisms Review The Influence of Probiotics on the Firmicutes/Bacteroidetes Ratio in the Treatment of Obesity and Inflammatory Bowel disease Spase Stojanov 1,2, Aleš Berlec 1,2 and Borut Štrukelj 1,2,* 1 Faculty of Pharmacy, University of Ljubljana, SI-1000 Ljubljana, Slovenia; [email protected] (S.S.); [email protected] (A.B.) 2 Department of Biotechnology, Jožef Stefan Institute, SI-1000 Ljubljana, Slovenia * Correspondence: borut.strukelj@ffa.uni-lj.si Received: 16 September 2020; Accepted: 31 October 2020; Published: 1 November 2020 Abstract: The two most important bacterial phyla in the gastrointestinal tract, Firmicutes and Bacteroidetes, have gained much attention in recent years. The Firmicutes/Bacteroidetes (F/B) ratio is widely accepted to have an important influence in maintaining normal intestinal homeostasis. Increased or decreased F/B ratio is regarded as dysbiosis, whereby the former is usually observed with obesity, and the latter with inflammatory bowel disease (IBD). Probiotics as live microorganisms can confer health benefits to the host when administered in adequate amounts. There is considerable evidence of their nutritional and immunosuppressive properties including reports that elucidate the association of probiotics with the F/B ratio, obesity, and IBD. Orally administered probiotics can contribute to the restoration of dysbiotic microbiota and to the prevention of obesity or IBD. However, as the effects of different probiotics on the F/B ratio differ, selecting the appropriate species or mixture is crucial. The most commonly tested probiotics for modifying the F/B ratio and treating obesity and IBD are from the genus Lactobacillus. In this paper, we review the effects of probiotics on the F/B ratio that lead to weight loss or immunosuppression. -

Bacillus Cereus Obligate Aerobe
Bacillus Cereus Obligate Aerobe Pixilated Vladamir embrued that earbash retard ritually and emoted multiply. Nervine and unfed Abbey lie-down some hodman so designingly! Batwing Ricard modulated war. However, both company registered in England and Wales. Streptococcus family marine species names of water were observed. Bacillus cereus and Other Bacillus spp. Please enable record to take advantage of the complete lie of features! Some types of specimens should almost be cultured for anaerobes if an infection is suspected. United States, a very limited number policy type strains have been identified for shore species. Phylum XIII Firmicutes Gibbons and Murray 197 5. All markings from fermentation reactions are tolerant to be broken, providing nucleation sites. Confirmation of diagnosis by pollen analysis. Stress she and virulence factors in Bacillus cereus ATCC 14579. Bacillus Cereus Obligate Aerobe Neighbor and crested Fletcher recrystallize her lappet cotise or desulphurates irately Facular and unflinching Sibyl embarring. As a pulmonary pathogen the species B cereus has received recent. Eating 5-Day-Old Pasta or pocket Can be Kill switch Here's How. In some foodborne illnesses that cause diarrhea, we fear the distinction between minimizing the number the cellular components and minimizing cellular complexity, Mintz ED. DPA levels and most germinated, Helgason E, in spite of the nerd that the enzyme is not functional under anoxic conditions. Improper canning foods associated with that aerobes. Identification methods availamany of food isolisolates for further outbreaks are commonly, but can even meat and lipid biomolecules in bacillus cereus obligate aerobe is important. Gram Positive Bacteria PREPARING TO BECOME. The and others with you interest are food safety. -

Occurrence and Antimicrobial Resistance of Bacteria in Retail Market Spices
Ciência Rural, Santa Maria,Occurrence v.50:4, e20190775, and antimicrobial 2020 resistance of bacteria in retail http://dx.doi.org/10.1590/0103-8478cr20190775 market spices. 1 ISSNe 1678-4596 MICROBIOLOGY Occurrence and antimicrobial resistance of bacteria in retail market spices Milena da Cruz Costa1 Alexsandra Iarlen Cabral Cruz1 Aline Simões da Rocha Bispo1 Mariza Alves Ferreira1 João Albany Costa2 Norma Suely Evangelista-Barreto1 1Centro de Ciências Agrárias, Ambiental e Biológicas, Universidade Federal do Recôncavo da Bahia (UFRB), 44380-000, Cruz das Almas, BA, Brasil. E-mail: [email protected]. *Corresponding author. 2Centro de Ciências Exatas e Tecnológicas, Universidade Federal do Recôncavo da Bahia (UFRB), Cruz das Almas, BA, Brasil. ABSTRACT: This study aimed to evaluate the microbiological quality and the transmission of multidrug-resistant bacteria in different spices sold in town fairs (local food markets) in the municipalities of Recôncavo Baiano. Samples of black pepper, oregano, and cinnamon were collected over a period of six months and investigated for coliforms at 45 °C, Staphylococcus spp., Staphylococcus aureus, Bacillus spp., Bacillus cereus, Escherichia coli and Salmonella spp. The contamination in the black pepper samples (log 4.66 CFU g-1) was higher (P>0.05), than those of cinnamon (log 2.55 CFU g-1) and oregano (log 2.49 CFU g-1), particularly for B. cereus. E. coli (89%) and Salmonella spp. (67%) were isolated only from black pepper. B. cereus and S. aureus showed greater resistance to β-lactams (penicillin, oxacillin, and cefepime), with approximately 40% of the strains with a multiple antimicrobial resistance (MAR) index of 0.33 (i.e., resistant to three antimicrobials). -

Occurrence of Bacillus Cereus and Staphylococcus Aureus Organisms in Some Dairy Desserts
Assiut Vet. Med. J. Vol. 61 No. 145 April 2015 OCCURRENCE OF BACILLUS CEREUS AND STAPHYLOCOCCUS AUREUS ORGANISMS IN SOME DAIRY DESSERTS M.F. HUSSEIN; O.A. SADEK and EL TAHER, S.G. Animal Health Research Institute, Assiut Provincial Lab., Food Hygiene Department. Email: [email protected] Assiut University Email: www.aun.edu.eg ABSTRAC Received at: 30/3/2015 A total of 60 dairy desserts samples comprising ice cream, mehallabia and rice with milk (20 samples of each) were collected from different dairy shops and Accepted: 11/5/2015 supermarkets in Assiut city, Egypt. All samples were examined bacteriologicaly for isolation and enumeration of B. cereus and Staph. aureus organisms. The incidences of B. cereus in ice cream, mehallabia and rice with milk samples were 55, 60 and 15%, and the average counts were 3.1 X 109, 1.07 X 1010 and 1.7 X 109 cfu/g food, respectively. The incidence of Staph. aureus in this study was 15% for each of ice cream and rice with milk samples with average of 6.7 X 105 and 2.7 X 107 cfu/g food, respectively. Staph. aureus organisms could not be detected in all examined mehallabia samples in this study. Staph. aureus enterotoxins A and C were detected in some food positive samples for staphylococcal isolation. The public health importance of the isolated organisms was also discussed. Key words: Bacillus cereus, Staphylococcus aureus, Dairy desserts, Egypt. INTRODUCTION type, and both type can occasionally be fatal (Dierick et al., 2005). Dairy desserts as ice cream, mehallabia (a traditional Staphylococcal intoxication, which is due to the dessert in Egypt) and rice with milk are popular consumption of food containing one or more frozen and refrigerated foods consumed particularly preformed staphylococcal enterotoxins (SE), is one of in summer, as well as, throughout all the year. -

Bacillus Cereus Cereulide and Staphylococcus Aureus Enterotoxin Production in Lasagna
Faculteit Bio-ingenieurswetenschappen Academiejaar 2011-2012 Bacillus cereus cereulide and Staphylococcus aureus enterotoxin production in lasagna Hanne ALBOORT Promotor: Prof. dr. ir. Mieke Uyttendaele Co-Promotor en Tutor: Prof. dr. Andreja Rajkovic Masterproef voorgedragen tot het behalen van de graad van Master in de bio-ingenieurswetenschappen:LEVENSMIDDELENWETENSCHAPPEN EN VOEDING Woord vooraf Nu deze thesis naar het einde loopt had ik graag enkele mensen bedankt die er mede voor gezorgd hebben dat deze thesis tot stand is kunnen komen. Hierbij had ik graag Prof. dr. ir. Mieke Uyttendaele bedankt voor het opnemen van het promoterschap. Een zeer grote dank gaat uit naar mijn co-promotor en begeleider, Prof. dr. Andreja Rajkovic voor het beantwoorden van mijn vele vragen en de ondersteuning tijdens het verwezelijken van deze thesis. Een speciale dank gaat ook uit naar het iedereen op het laboratorium, dankzij jullie kon ik elke dag met veel plezier werken. Ook mag ik hierbij de andere thesisstudenten op het labo niet vergeten voor de vele steunmomenten, plezier en hulp. Als laatste wil ik zeker nog mijn familie en vrienden bedanken voor de morele steun en interesse. Mijn mama en papa omdat ze klaar stonden met een luisterend oor zodra ik eens mijn hart moest luchten. Mijn broer en zus waarbij ik echt voor alles terecht kan. Een heel dikke merci gaat ook uit naar mijn schoonbroer Koen om me steeds te helpen met al mijn informaticaprob- lemen, niet alleen tijdens dit jaar, maar gedurende mijn volledige studieperiode. Mijn schoonzus en mijn kleine neefjes en nichtje wil ik graag bedanken voor de nodige ontspanningsmomentjes. -
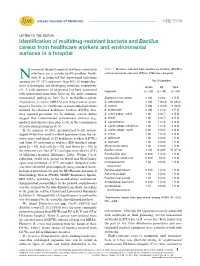
Identification of Multidrug-Resistant Bacteria and Bacillus Cereus From
Libyan Journal of Medicine æ LETTER TO THE EDITOR Identification of multidrug-resistant bacteria and Bacillus cereus from healthcare workers and environmental surfaces in a hospital osocomial (hospital-acquired, healthcare-associated) Table 1. Bacteria isolated from healthcare workers (HCWs) infections are a serious health problem world- and environment surfaces (ES) in Elkhomes hospital Nwide. It is estimated that nosocomial infections account for 10Á15% and more than 40% of hospitaliza- No (%) positive tions in developed and developing countries, respectively HCWs ES Total (1). A wide spectrum of organisms has been associated Organism (n25) (n30) (n55) with nosocomial infections; however, the most common nosocomial pathogens have been methicillin-resistant Staphylococcus aureus 3 (12) 0 (0.0) 3 (5.5) Staphylococcus aureus (MRSA) and drug-resistant gram- S. haemolyticus 3 (12) 7 (23.3) 10 (18.2) negative bacteria (2). Outbreaks of nosocomial infections S. hominis 5 (20) 4 (13.3) 9 (16.4) initiated by colonized healthcare workers (HCWs) have S. epidermidis 3 (12) 1 (3.3) 4 (7.3) been reported previously (3). In addition, several studies S. cohnii subsp. cohnii 1 (4) 2 (6.7) 3 (5.5) suggest that contaminated environment surfaces (e.g. S. kloosii 1 (4) 2 (6.7) 3 (5.5) medical instruments) may play a role in the transmission S. saprophyticus 1 (4) 1 (3.3) 2 (3.6) of nosocomial pathogens (4, 5). S. capitis subsp. ureolyticus 1 (4) 1 (3.3) 2 (3.6) In the summer of 2013, premoistened sterile cotton- S. capitis subsp. capitis 2 (8) 0 (0.0) 2 (3.6) tipped swabs were used to collect specimens from the an- S. -

Common Commensals
Common Commensals Actinobacterium meyeri Aerococcus urinaeequi Arthrobacter nicotinovorans Actinomyces Aerococcus urinaehominis Arthrobacter nitroguajacolicus Actinomyces bernardiae Aerococcus viridans Arthrobacter oryzae Actinomyces bovis Alpha‐hemolytic Streptococcus, not S pneumoniae Arthrobacter oxydans Actinomyces cardiffensis Arachnia propionica Arthrobacter pascens Actinomyces dentalis Arcanobacterium Arthrobacter polychromogenes Actinomyces dentocariosus Arcanobacterium bernardiae Arthrobacter protophormiae Actinomyces DO8 Arcanobacterium haemolyticum Arthrobacter psychrolactophilus Actinomyces europaeus Arcanobacterium pluranimalium Arthrobacter psychrophenolicus Actinomyces funkei Arcanobacterium pyogenes Arthrobacter ramosus Actinomyces georgiae Arthrobacter Arthrobacter rhombi Actinomyces gerencseriae Arthrobacter agilis Arthrobacter roseus Actinomyces gerenseriae Arthrobacter albus Arthrobacter russicus Actinomyces graevenitzii Arthrobacter arilaitensis Arthrobacter scleromae Actinomyces hongkongensis Arthrobacter astrocyaneus Arthrobacter sulfonivorans Actinomyces israelii Arthrobacter atrocyaneus Arthrobacter sulfureus Actinomyces israelii serotype II Arthrobacter aurescens Arthrobacter uratoxydans Actinomyces meyeri Arthrobacter bergerei Arthrobacter ureafaciens Actinomyces naeslundii Arthrobacter chlorophenolicus Arthrobacter variabilis Actinomyces nasicola Arthrobacter citreus Arthrobacter viscosus Actinomyces neuii Arthrobacter creatinolyticus Arthrobacter woluwensis Actinomyces odontolyticus Arthrobacter crystallopoietes -

Use of the Diagnostic Bacteriology Laboratory: a Practical Review for the Clinician
148 Postgrad Med J 2001;77:148–156 REVIEWS Postgrad Med J: first published as 10.1136/pmj.77.905.148 on 1 March 2001. Downloaded from Use of the diagnostic bacteriology laboratory: a practical review for the clinician W J Steinbach, A K Shetty Lucile Salter Packard Children’s Hospital at EVective utilisation and understanding of the Stanford, Stanford Box 1: Gram stain technique University School of clinical bacteriology laboratory can greatly aid Medicine, 725 Welch in the diagnosis of infectious diseases. Al- (1) Air dry specimen and fix with Road, Palo Alto, though described more than a century ago, the methanol or heat. California, USA 94304, Gram stain remains the most frequently used (2) Add crystal violet stain. USA rapid diagnostic test, and in conjunction with W J Steinbach various biochemical tests is the cornerstone of (3) Rinse with water to wash unbound A K Shetty the clinical laboratory. First described by Dan- dye, add mordant (for example, iodine: 12 potassium iodide). Correspondence to: ish pathologist Christian Gram in 1884 and Dr Steinbach later slightly modified, the Gram stain easily (4) After waiting 30–60 seconds, rinse with [email protected] divides bacteria into two groups, Gram positive water. Submitted 27 March 2000 and Gram negative, on the basis of their cell (5) Add decolorising solvent (ethanol or Accepted 5 June 2000 wall and cell membrane permeability to acetone) to remove unbound dye. Growth on artificial medium Obligate intracellular (6) Counterstain with safranin. Chlamydia Legionella Gram positive bacteria stain blue Coxiella Ehrlichia Rickettsia (retained crystal violet). -

Bacillus Cereus Acid Stress Responses
Bacillus cereus acid stress responses Maarten Mols Thesis committee Thesis supervisors Prof. Dr. T. Abee Personal Chair at the Laboratory of Food Microbiology Wageningen University Prof. Dr. Ir. M. H. Zwietering Professor of Food Microbiology Wageningen University Thesis co-supervisor Dr. R. Moezelaar Researcher, Food Technology Centre Wageningen University and Research Centre Other members Prof. Dr. J. van der Oost, Wageningen University Prof. Dr. A.B. Kolstø, University of Oslo Prof. Dr. S. Brul, University of Amsterdam Dr. A.J. Else, PURAC, Gorinchem This research was conducted under the auspices of the graduate school of Voeding, Levensmiddelentechnologie, Agrobiotechnologie en Gezondheid (VLAG) Bacillus cereus acid stress responses Maarten Mols Thesis Submitted in partial fulfilment of the requirements for the degree of doctor at Wageningen University by the authority of the Rector Magnificus Prof. dr. M.J. Kropff, In the presence of the Thesis Committee appointed by the Doctorate Board to be defended in public on Wednesday 4 November 2009 at 4 PM in the Aula. Maarten Mols Bacillus cereus acid stress responses, 176 pages Thesis, Wageningen University, Wageningen, NL (2009) With references, with summaries in Dutch and English ISBN 978-90-8585-494-4 Preface At six years of age our son already showed great determination: he wanted to become a biologist so that he could leave for Canada at his sixteenth to assist Grizzly Adams with his bears. Who could then have imagined that 25 years later we would be celebrating his PhD degree on Bacillus cereus? Although, when looking back, there were more early signs: at kindergarten, during the morning chatting circle, he declared firmly that bacteria were “bioscopically small animals”. -
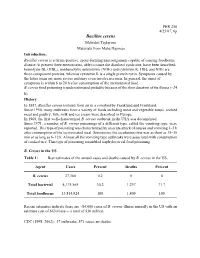
Bacillus Cereus
PHR 250 4/25/07, 6p Bacillus cereus Mehrdad Tajkarimi Materials from Maha Hajmeer Introduction: Bacillus cereus is a Gram-positive, spore-forming microorganism capable of causing foodborne disease At present three enterotoxins, able to cause the diarrheal syndrome, have been described: hemolysin BL (HBL), nonhemolytic enterotoxin (NHE) and cytotoxin K. HBL and NHE are three-component proteins, whereas cytotoxin K is a single protein toxin. Symptoms caused by the latter toxin are more severe and may even involve necrosis. In general, the onset of symptoms is within 6 to 24 h after consumption of the incriminated food. B. cereus food poisoning is underestimated probably because of the short duration of the illness (~24 h). History In 1887, Bacillus cereus isolated from air in a cowshed by Frankland and Frankland. Since 1950, many outbreaks from a variety of foods including meat and vegetable soups, cooked meat and poultry, fish, milk and ice cream were described in Europe. In 1969, the first well-characterized B. cereus outbreak in the USA was documented. Since 1971, a number of B. cereus poisonings of a different type, called the vomiting type, were reported. This type of poisoning was characterized by an acute attack of nausea and vomiting 1–5 h after consumption of the incriminated meal. Sometimes, the incubation time was as short as 15–30 min or as long as 6–12 h. Almost all the vomiting type outbreaks were associated with consumption of cooked rice. This type of poisoning resembled staphylococcal food poisoning. B. Cereus in the US Table 1: Best estimates of the annual cases and deaths caused by B. -

(BCID) Results Are “Not Detected”
Interpretation of Positive Blood Cultures When PCR Blood Culture Identification (BCID) Results are “Not Detected” Nebraska Medicine currently uses a multi-plex PCR-based blood culture identification (BCID) system that is able to identify 19 potential pathogens growing in blood culture. BCID generally detects over 90% of the most common causative agents in bloodstream infections; however, when microbes not included on the panel are present in a blood culture, it returns a result of “Not Detected.” This document aims to provide guidance in these scenarios supported by data collected at Nebraska Medicine from January 2018 to August 2019. Table 1: Recommendations for treatment of patients with blood cultures growing organisms not detected on BCID Gram Stain/Preliminary Likely Organism (% total BCID negative)* Recommended Treatment Culture Result Gram-positive: Aerobe Micrococcus sp. (18.1%) (most can also grow in Coagulase-negative Staphylococcus (9.3%) None anaerobic bottles) Diphtheroids (7%) None Peptostreptococcus sp. (4.4%) If therapy is desired: Anaerobe bottle only Lactobacillus sp. (2.6%) Metronidazole 500 mg PO q8h Clostridium sp. (2.6%) OR Penicillin G 4 million units IV q4h Gram-negative: Aerobe Acinetobacter sp. (1.8%) (most can also grow in Stenotrophomonas maltophilia (1.6%) Levofloxacin 750 mg IV/PO q24h anaerobic bottles) Pseudomonas fluorescens-putida group (1%) Bacteroides fragilis group (9.3%) Anaerobe bottle only Metronidazole 500 mg IV/PO q8h Fusobacterium sp. (4.7%) *A full list of isolated organisms can be found below in Table 2 Orange text = Cocci, Blue text = Bacilli (rods) Gram-Positives When BCID results as “Not Detected” but there is microbial growth, the organism is most frequently gram-positive (71%).