Hurricane Ivan
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

"National List of Vascular Plant Species That Occur in Wetlands: 1996 National Summary."
Intro 1996 National List of Vascular Plant Species That Occur in Wetlands The Fish and Wildlife Service has prepared a National List of Vascular Plant Species That Occur in Wetlands: 1996 National Summary (1996 National List). The 1996 National List is a draft revision of the National List of Plant Species That Occur in Wetlands: 1988 National Summary (Reed 1988) (1988 National List). The 1996 National List is provided to encourage additional public review and comments on the draft regional wetland indicator assignments. The 1996 National List reflects a significant amount of new information that has become available since 1988 on the wetland affinity of vascular plants. This new information has resulted from the extensive use of the 1988 National List in the field by individuals involved in wetland and other resource inventories, wetland identification and delineation, and wetland research. Interim Regional Interagency Review Panel (Regional Panel) changes in indicator status as well as additions and deletions to the 1988 National List were documented in Regional supplements. The National List was originally developed as an appendix to the Classification of Wetlands and Deepwater Habitats of the United States (Cowardin et al.1979) to aid in the consistent application of this classification system for wetlands in the field.. The 1996 National List also was developed to aid in determining the presence of hydrophytic vegetation in the Clean Water Act Section 404 wetland regulatory program and in the implementation of the swampbuster provisions of the Food Security Act. While not required by law or regulation, the Fish and Wildlife Service is making the 1996 National List available for review and comment. -

Cocoa Beach Maritime Hammock Preserve Management Plan
MANAGEMENT PLAN Cocoa Beach’s Maritime Hammock Preserve City of Cocoa Beach, Florida Florida Communities Trust Project No. 03 – 035 –FF3 Adopted March 18, 2004 TABLE OF CONTENTS SECTION PAGE I. Introduction ……………………………………………………………. 1 II. Purpose …………………………………………………………….……. 2 a. Future Uses ………….………………………………….…….…… 2 b. Management Objectives ………………………………………….... 2 c. Major Comprehensive Plan Directives ………………………..….... 2 III. Site Development and Improvement ………………………………… 3 a. Existing Physical Improvements ……….…………………………. 3 b. Proposed Physical Improvements…………………………………… 3 c. Wetland Buffer ………...………….………………………………… 4 d. Acknowledgment Sign …………………………………..………… 4 e. Parking ………………………….………………………………… 5 f. Stormwater Facilities …………….………………………………… 5 g. Hazard Mitigation ………………………………………………… 5 h. Permits ………………………….………………………………… 5 i. Easements, Concessions, and Leases …………………………..… 5 IV. Natural Resources ……………………………………………..……… 6 a. Natural Communities ………………………..……………………. 6 b. Listed Animal Species ………………………….…………….……. 7 c. Listed Plant Species …………………………..…………………... 8 d. Inventory of the Natural Communities ………………..………….... 10 e. Water Quality …………..………………………….…..…………... 10 f. Unique Geological Features ………………………………………. 10 g. Trail Network ………………………………….…..………..……... 10 h. Greenways ………………………………….…..……………..……. 11 i Adopted March 18, 2004 V. Resources Enhancement …………………………..…………………… 11 a. Upland Restoration ………………………..………………………. 11 b. Wetland Restoration ………………………….…………….………. 13 c. Invasive Exotic Plants …………………………..…………………... 13 d. Feral -
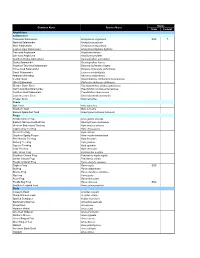
St. Joseph Bay Native Species List
Status Common Name Species Name State Federal Amphibians Salamanders Flatwoods Salamander Ambystoma cingulatum SSC T Marbled Salamander Ambystoma opacum Mole Salamander Ambystoma talpoideum Eastern Tiger Salamander Ambystoma tigrinum tigrinum Two-toed Amphiuma Amphiuma means One-toed Amphiuma Amphiuma pholeter Southern Dusky Salamander Desmognathus auriculatus Dusky Salamander Desmognathus fuscus Southern Two-lined Salamander Eurycea bislineata cirrigera Three-lined Salamander Eurycea longicauda guttolineata Dwarf Salamander Eurycea quadridigitata Alabama Waterdog Necturus alabamensis Central Newt Notophthalmus viridescens louisianensis Slimy Salamander Plethodon glutinosus glutinosus Slender Dwarf Siren Pseudobranchus striatus spheniscus Gulf Coast Mud Salamander Pseudotriton montanus flavissimus Southern Red Salamander Pseudotriton ruber vioscai Eastern Lesser Siren Siren intermedia intermedia Greater Siren Siren lacertina Toads Oak Toad Bufo quercicus Southern Toad Bufo terrestris Eastern Spadefoot Toad Scaphiopus holbrooki holbrooki Frogs Florida Cricket Frog Acris gryllus dorsalis Eastern Narrow-mouthed Frog Gastrophryne carolinensis Western Bird-voiced Treefrog Hyla avivoca avivoca Cope's Gray Treefrog Hyla chrysoscelis Green Treefrog Hyla cinerea Southern Spring Peeper Hyla crucifer bartramiana Pine Woods Treefrog Hyla femoralis Barking Treefrog Hyla gratiosa Squirrel Treefrog Hyla squirella Gray Treefrog Hyla versicolor Little Grass Frog Limnaoedus ocularis Southern Chorus Frog Pseudacris nigrita nigrita Ornate Chorus Frog Pseudacris -

Hydrologic Restoration Plan for the Tate's Hell State Forest
Tate’s Hell State Forest Hydrologic Restoration Plan Volume I Northwest Florida Water Management District 81 Water Management Drive, Havana, Florida 32333-4712 Florida Division of Forestry 290 Airport Road, Carrabelle, FL 32322 i GOVERNING BOARD George Roberts, Chair Panama City Philip McMillan, Vice Chair Blountstown Steve Ghazvini, Secretary/Treasurer Tallahassee Stephanie Bloyd Joyce Estes Jerry Pate Panama City Beach Eastpoint Pensacola Peter Antonacci Ralph Rish Tim Norris Tallahassee Port St. Joe Santa Rosa Beach Douglas E. Barr Executive Director For additional information, write or call: Northwest Florida Water Management District 81 Water Management Drive Havana, Florida 32333-4712 (850) 539-5999 DISTRICT OFFICES Headquarters 81 Water Management Drive Havana, FL 32333-4712 Telephone (850) 539-5999 Fax (850) 539-2777 Crestview 800 Hospital Drive Crestview, Florida 32539-7385 Telephone (850) 683-5044 Fax (850) 683-5050 Tallahassee The Delaney Center Building, Suite 2-D 2252 Killearn Center Boulevard Tallahassee, FL 32309-3573 Telephone (850) 921-2986 Fax (850) 921-3083 Pensacola (Field Office) 2261 West Nine Mile Road Pensacola, FL 32534-9416 Telephone (850) 484-5125 Fax (850) 484-5133 Marianna 4765 Pelt Street Marianna, FL 32446-6846 Telephone (850) 482-9522 Fax (850) 482-1376 Econfina (Field Office) 6418 E. Highway 20 Youngstown, FL 32466-3808 Telephone (850) 722-9919 Fax (850) 722-8982 Acknowledgements: This Hydrologic Restoration Plan is the result of the work and contributions of many individuals. At the Northwest Florida Water Management District, Ron Bartel, Director of the Resource Management Division, supervised this effort and provided oversight during the plan development process. We gratefully acknowledge Linda Chaisson, who participated in many of the field reviews along with Nick Wooten, Duncan Cairns, David Clayton, and John Crowe, who provided valuable guidance and suggestions regarding the plan‟s content. -

Mississippi Natural Heritage Program Special Plants - Tracking List -2018
MISSISSIPPI NATURAL HERITAGE PROGRAM SPECIAL PLANTS - TRACKING LIST -2018- Approximately 3300 species of vascular plants (fern, gymnosperms, and angiosperms), and numerous non-vascular plants may be found in Mississippi. Many of these are quite common. Some, however, are known or suspected to occur in low numbers; these are designated as species of special concern, and are listed below. There are 495 special concern plants, which include 4 non- vascular plants, 28 ferns and fern allies, 4 gymnosperms, and 459 angiosperms 244 dicots and 215 monocots. An additional 100 species are designated “watch” status (see “Special Plants - Watch List”) with the potential of becoming species of special concern and include 2 fern and fern allies, 54 dicots and 44 monocots. This list is designated for the primary purposes of : 1) in environmental assessments, “flagging” of sensitive species that may be negatively affected by proposed actions; 2) determination of protection priorities of natural areas that contain such species; and 3) determination of priorities of inventory and protection for these plants, including the proposed listing of species for federal protection. GLOBAL STATE FEDERAL SPECIES NAME COMMON NAME RANK RANK STATUS BRYOPSIDA Callicladium haldanianum Callicladium Moss G5 SNR Leptobryum pyriforme Leptobryum Moss G5 SNR Rhodobryum roseum Rose Moss G5 S1? Trachyxiphium heteroicum Trachyxiphium Moss G2? S1? EQUISETOPSIDA Equisetum arvense Field Horsetail G5 S1S2 FILICOPSIDA Adiantum capillus-veneris Southern Maidenhair-fern G5 S2 Asplenium -

BIOLOGICAL Evaluation Analysis Uint 23 9/13/2010
BIOLOGICAL Evaluation Analysis Uint 23 9/13/2010 BIOLOGICAL EVALUATION OF FOREST MANAGEMENT ACTIVITIES PROPOSED FOR ANALYSIS UNIT 23 (Compartments 253, 254, 255, 256, 257 and 259) Gloster and Crosby Quadrangles USDA Forest Service Southern Region (8) National Forests in Mississippi Homochitto National Forest Homochitto Ranger District Mississippi Prepared by: Kenneth L. Gordon _____________________________ Date____________________ Wildlife Biologist, USDA Forest Service Concurred by: KATHY W. LUNCEFORD _____________________ Date____________________ Environmental Coordinator for Mississippi, US Fish and Wildlife Service Homochitto Ranger District Page 1 BIOLOGICAL Evaluation Analysis Uint 23 9/13/2010 Introduction This Biological Evaluation (BE) documents the likely impacts on proposed, endangered, threatened, and sensitive (TES) species from forest management activities proposed for Analysis Unit 23. AU 23 is located in Amite and Wilkinson counties, Mississippi and is part of the Foster Creek subwatershed in the Homochitto River basin.. As a result of a recent court decision, Forest Plan Amendment # 16 and the Region 8 Supplement to FSM 2670 are no longer in effect. This BE follows the process used to decide when to inventory for TES species that is consistent with the requirement found in the Vegetation Management EIS for the Coastal Plain and Piedmont. This BE is in accordance with direction given in Forest Service Manual (FSM) 2672 to meet the 1989 Vegetation Management standard. As part of the NEPA decision making process, the BE provides a review of Forest Service (FS) activities in sufficient detail to determine how an action will affect any TES species. TES species, taken from both state and federal lists, are species whose viability is most likely to be put at risk from management actions. -

The Draft Genome of Ruellia Speciosa (Beautiful Wild Petunia: Acanthaceae) Yongbin Zhuang1,2 and Erin A
Dna Research, 2017, 24(2), 179–192 doi: 10.1093/dnares/dsw054 Advance Access Publication Date: 27 January 2017 Full Paper Full Paper The draft genome of Ruellia speciosa (Beautiful Wild Petunia: Acanthaceae) Yongbin Zhuang1,2 and Erin A. Tripp1,2,* 1Department of Ecology and Evolutionary Biology, University of Colorado, UCB 334, Boulder, CO 80309, USA and 2Museum of Natural History, University of Colorado, UCB 350, Boulder, CO 80309, USA *To whom correspondence should be addressed. Email: [email protected] Edited by Dr. Sachiko Isobe Received 18 July 2016; Editorial decision 16 November 2016; Accepted 17 November 2016 Abstract The genus Ruellia (Wild Petunias; Acanthaceae) is characterized by an enormous diversity of floral shapes and colours manifested among closely related species. Using Illumina platform, we reconstructed the draft genome of Ruellia speciosa, with a scaffold size of 1,021 Mb (or 1.02 Gb) and an N50 size of 17,908 bp, spanning 93% of the estimated genome (1.1 Gb). The draft assembly predicted 40,124 gene models and phylogenetic analyses of four key en- zymes involved in anthocyanin colour production [flavanone 3-hydroxylase (F3H), flavonoid 30- hydroxylase (F30H), flavonoid 30,50-hydroxylase (F3050H), and dihydroflavonol 4-reductase (DFR)] found that most angiosperms here sampled harboured at least one copy of F3H, F30H, and DFR. In contrast, fewer than one-half (but including R. speciosa) harboured a copy of F3050H, support- ing observations that blue flowers and/or fruits, which this enzyme is required for, are less com- mon among flowering plants. Ka/Ks analyses of duplicated copies of F30H and DFR in R. -
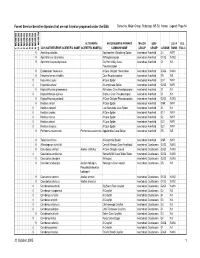
Sensitive Species That Are Not Listed Or Proposed Under the ESA Sorted By: Major Group, Subgroup, NS Sci
Forest Service Sensitive Species that are not listed or proposed under the ESA Sorted by: Major Group, Subgroup, NS Sci. Name; Legend: Page 94 REGION 10 REGION 1 REGION 2 REGION 3 REGION 4 REGION 5 REGION 6 REGION 8 REGION 9 ALTERNATE NATURESERVE PRIMARY MAJOR SUB- U.S. N U.S. 2005 NATURESERVE SCIENTIFIC NAME SCIENTIFIC NAME(S) COMMON NAME GROUP GROUP G RANK RANK ESA C 9 Anahita punctulata Southeastern Wandering Spider Invertebrate Arachnid G4 NNR 9 Apochthonius indianensis A Pseudoscorpion Invertebrate Arachnid G1G2 N1N2 9 Apochthonius paucispinosus Dry Fork Valley Cave Invertebrate Arachnid G1 N1 Pseudoscorpion 9 Erebomaster flavescens A Cave Obligate Harvestman Invertebrate Arachnid G3G4 N3N4 9 Hesperochernes mirabilis Cave Psuedoscorpion Invertebrate Arachnid G5 N5 8 Hypochilus coylei A Cave Spider Invertebrate Arachnid G3? NNR 8 Hypochilus sheari A Lampshade Spider Invertebrate Arachnid G2G3 NNR 9 Kleptochthonius griseomanus An Indiana Cave Pseudoscorpion Invertebrate Arachnid G1 N1 8 Kleptochthonius orpheus Orpheus Cave Pseudoscorpion Invertebrate Arachnid G1 N1 9 Kleptochthonius packardi A Cave Obligate Pseudoscorpion Invertebrate Arachnid G2G3 N2N3 9 Nesticus carteri A Cave Spider Invertebrate Arachnid GNR NNR 8 Nesticus cooperi Lost Nantahala Cave Spider Invertebrate Arachnid G1 N1 8 Nesticus crosbyi A Cave Spider Invertebrate Arachnid G1? NNR 8 Nesticus mimus A Cave Spider Invertebrate Arachnid G2 NNR 8 Nesticus sheari A Cave Spider Invertebrate Arachnid G2? NNR 8 Nesticus silvanus A Cave Spider Invertebrate Arachnid G2? NNR -

Field Trip to Apalachicola National Forest, Florida Panhandle, South
Volume 88 Number 6 November 2013 Georgia Botanical Society Field Trip to Apalachicola National Forest, IN THIS Florida Panhandle, South Georgia ISSUE: Photography August 17-18, 2013 Text by Virginia Craig and Jim Drake Project - P2 The 6:00 A.M. Saturday downpour seemed to validate the forecasted weekend torrents. The day’s plan was to roadside botanize in the Apalachicola National Waycross FT Forest (ANF) at several stops along Florida Highway 65 and adjacent roads. - P6 Our intrepid group departed the Quincy, Florida Hampton Inn at around 9:15 2014 A.M. for an approximately 150 mile foray into the ANF in Liberty and Franklin Pilgrimage— Counties – so far, so good, very cloudy, rain holding off. At Hosford, Florida, we turned onto Hwy 65 going southwest. At the first stop, we found a few stems of P10 yellow crested orchid (Platanthera cristata) – past prime. These plants were Upcoming almost finished blooming as they are the first of the Platanthera in this area to Field Trips - bloom. Near the next stop we began looking for Catesby's lily (Lilium catesbaei). After spotting a few blooms, we pulled off and found several more P11 flowers a little further down the road. Lilium catesbaei (above) by Virginia Craig. Platanthera ciliaris (right) by Ed McDowell Continued on page 3 2 BotSoc News, November 2013 The Botanical Plight in Georgia Have you ever bemoaned the fact that you could not find good or useful plant pictures on the internet or that you did not have an “expert” to help you identify a plant? Read on – I may have an answer for you. -

Draft Environmental Assessment
United States Department of Environmental Assessment Agriculture Forest Mechanical Fuel Reduction Service August 2009 PALS No. 28526 Apalachicola and Wakulla Ranger Districts, Apalachicola National Forest, Liberty and Wakulla Counties, Florida Deciding Officer: Marcus A. Beard District Ranger For Information Contact: Susan Fitzgerald Apalachicola Ranger District PO Box 579 Bristol, FL 32321 The U.S. Department of Agriculture (USDA) prohibits discrimination in all its programs and activities on the basis of race, color, national origin, gender, religion. age, disability, political beliefs, sexual orientation, or marital or family status. (Not all prohibited bases apply to all programs.) Persons with disabilities who require alternative means for communication of program information (Braille, large print, audiotape, etc.) should contact USDA's TARGET Center at (202) 720- 2600 (voice and TDD). To file a complaint of discrimination, write USDA, Director, Office of Civil Rights, Room 326-W, Whitten Building, 14th and Independence Avenue, SW, Washington, DC 20250-9410 or call (202) 720-5964 (voice and TDD). USDA is an equal opportunity provider and employer. Table of Contents Page SUMMARY i INTRODUCTION 1 Background 1 Purpose and Need for Action 1 Proposed Action 2 Decision Framework 3 Public Involvement 3 Issues 4 ALTERNATIVES 4 Alternatives 4 Coordination Measures 5 Comparison of Alternatives 5 ENVIRONMENTAL CONSEQUENCES 6 Physical Components 6 Air, Soil, and Water 6 Biological Components 8 Vegetation 8 Plant Communities 8 Management Indicator -

Rare Vascular Plant Taxa Associated with the Longleaf Pine Ecosystems: Patterns in Taxonomy and Ecology
Rare Vascular Plant Taxa Associated with the Longleaf Pine Ecosystems: Patterns in Taxonomy and Ecology Joan Walker U.S.D.A. Forest Service, Southeastern Forest Experiment Station, Department of Forest Resources, Clemson University, Clemson, SC 29634 ABSTRACT Ecological, taxonomic and biogeographical characteristics are used to describe the group of 187 rare vascular plant taxa associated with longleaf pine (Pinus palustris) throughout its range. Taxonomic and growth form distributions mirror the patterns of common plus rare taxa in the flora. Most of the species have rather narrow habitat preferences, and narrow geo graphic ranges, but a few rare sp~cies with broad habitat tolerances and wider geographic ranges are identified. Ninety-six local endemics are associated with longleaf pine ecosystems. This incidence is as high as in other comparably-sized endemic-rich areas in North America. A distinct geographic trend in rare species composition is indicated. Species fall into 4 groups: Florida longleaf associates, south Atlantic coastal plain, east Gulf coastal plain, and west Gulf coastal plain species. Distributional factors that produce rarity must be considered in the development of conser vation strategies. Overall, conserving longleaf communities rangewide will protect .large ~ numbers of rare plant taxa in Southeastern United States. INTRODUCTION 1986), and inevitably the strategies required to con serve them will differ. Recently Hardin and White (1989) effectively focused conservationists' attentions on the high The purposes of this study are to (1) identify numbers of rare species associated with wiregrass the rare species associated with longleaf pine eco (Aristida stricta), a grass that dominates the ground systems rangewide; (2) characterize the rare spe layer of longleaf communities through a large part cies taxonomically and ecologically, in order to of its range, and over a broad range of longleaf identify patterns that may distinguish this group habitats. -

Alabama Inventory List
Alabama Inventory List The Rare, Threatened, & Endangered Plants & Animals of Alabama June 2004 Table of Contents INTRODUCTION .....................................................................................................................................................................1 DEFINITION OF HERITAGE RANKS .................................................................................................................................3 DEFINITIONS OF FEDERAL & STATE LISTED SPECIES STATUS.............................................................................5 AMPHIBIANS............................................................................................................................................................................6 BIRDS .........................................................................................................................................................................................7 MAMMALS...............................................................................................................................................................................10 FISHES.....................................................................................................................................................................................12 REPTILES ................................................................................................................................................................................16 CLAMS & MUSSELS ..............................................................................................................................................................18