Determination of the Reference Genes for Qrt-PCR Normalization and Expression Levels of MAT Genes Under Various Conditions in Ulocladium
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Protection Against Fungi in the Marketing of Grains and Byproducts
Protection against fungi in the marketing of grains and byproducts Ing. Agr. Juan M. Hernandez Vieyra ARGENT EXPORT S.A. May 2nd 2011 OBJECTIVE: To supply tools to eliminate fungus and bacteria contamination in maize and soybeans: Particularly: Stenocarpella maydis Cercospora sojina 2 • Powerfull Disinfectant of great efficacy in fungus, bacteria and virus • Produced by ICA Laboratories, South Africa. • aka SPOREKILL, VIRUKILL • Registered in more than 20 countries: USA, Australia, New Zeland, Brazil, Philipines, Israel • Product scientifically and field proven, with more than 15 years in the international market. • Registered at SENASA • Certifications: ISO 9001, GMP. 3 Properties of Sportek: – Based on a novel and patented quaternary amonio compound sintesis : didecil dimetil amonium chloride. – Excellent biodegradability thus, low environmental impact. – Really non corrosive and non oxidative. – Non toxic at recommended dosis . – Minimum inhibition concentration has a very low toxicity, LD 50>4000mg/Kg., lower than table salt. – High content of surfactants with excellent wetting capacity and penetration. – High efficacy in presence of organic matter, also with hard waters and heavy soils. – Non dependent of pH and is effective under a wide range of temperatures. 4 What is Sportek used for: To disinfect a wide spectrum of surfaces and feeds against: • Virus, • Bacteria, • Mycoplasma, • yeast, • Algae, • Fungus. 5 Where Sportek has been proven: VIRUKILL ES EFECTIVO CONTRA LOS VIRUS DE AVICULTURA, BACTERIAS HONGOS Y GRUPOS DE FAMILIA DE MICOPLASMA Hongos, levadura y EJEMPLOS DE VIRUS EJEMPLOS DE BACTERIAS ejemplos de Grupos de familia Ejemplos de Acinetobacter Ornithobacterium micoplasma patógenos anitratus rhinotracheale Birnaviridae Gumboro (IBD) Bacillus subtilis Pasteurella spores multocida Caliciviridae Feline calicivirus Bacilillus subtilis Pasteurella Aspergillus Níger vegetative volantium Coronaviridae Infectious bronchitis Bordatella spp. -

43-09-405 and 407 Final Combine
Pak. J. Bot ., 44(6): 2093-2102, 2012. A NEW SPECIES OF TIAROSPORELLA AZADARICHTA AND NEW FUNGAL RECORDS ON AZADIRACHTA INDICA FROM PAKISTAN SYED QAISER ABBAS 1, TEHREEMA IFTIKHAR 1* , MUBASHIR NIAZ 1, IFTIKHAR ALI 1, NABILA IFTIKHAR 1 AND ALIA ABBAS 2 1Department of Botany, Government College University, Alama Iqbal Road, Faisalabad, Pakistan 2Department of Botany, Federal Urdu University, Karachi, Pakistan *Corresponding author’s e-mail: [email protected] Abstract A new species of Tiarosporella azadarichta has been described on Azadirachta indica and some new fungal records viz ., Diplozythiella bambusina , Ulocladium chartarum , Cladosporium nigrellum, Cladosporium oxysporum. Didymostilbe coffeae , Muellerella pygmaea, Lasiodiplodia paraphysaria, Monochaetinula terminalae, Trimmatostroma sp., and Epidermophyton floccosum are reported on Azadirachta indica for the first time from Pakistan. Introduction reason a detailed survey of the plant for fungi was under taken in this study. Azadirachta indica (as Azadirachta Juss, Melia azadirachta ) belong to the family Meliaceae. Neem is its Materials and Methods common name. It is important due to its commercial and medicinal value. Azadirachta indica is also an important Samples of Azadirachta indica were collected from plant for its potential of anti-fungal, anti-bacterial and the different areas of District Faisalabad and Gojra. The insecticidal activity. Decline of trees due to fungi are different areas were G.C. University. Faisalabad, increasing tremendously in Pakistan especially in Punjab Agriculture University Faisalabad, Gutwala forest (park) and Sindh (Javed et al ., 2004; Khanzada et al ., 2011; Fateh Faisalabad, Tandlianwala and Gojra City. Methods and et al., 2011; Rheman et al., 2011; Farooq et al., 2011). materials are the same as described Abbas et al ., (2010). -

An Unusual Haemoid Fungi: Ulocladium, As a Cause Of
Volume 2 Number 2 (June 2010) 95-97 CASE Report An unusual phaeoid fungi: Ulocladium, as a cause of chronic allergic fungal sinusitis Kaur R, Wadhwa A1,Gulati A2, Agrawal AK2 1Department of Microbiology. 2Department of Otorhinolaryngology, Maulana Azad Medical College, New Delhi, India. Received: April 2010, Accepted: May 2010. ABSTRACT Allergic fungal sinusitis (AFS) has been recognized as an important cause of chronic sinusitis commonly caused by Aspergillus spp. and various dematiaceous fungi like Bipolaris, Alternaria, Curvalaria, and etc. Ulocladium botrytis is a non pathogenic environmental dematiaceous fungi, which has been recently described as a human pathogen. Ulocladium has never been associated with allergic fungal sinusitis but it was identified as an etiological agent of AFS in a 35 year old immunocompetent female patient presenting with chronic nasal obstruction of several months duration to our hospital. The patient underwent FESS and the excised polyps revealed Ulocladium as the causative fungal agent. Keywords: Ulocladium, Phaeoid Fungi, Chronic Sinusitis. INTRODUCTION CASE REPORT Chronic allergic sinusitis is a common condition A 35 year old female patient presented to the ENT responsible for the development of nasal polyps, out patient department of the Lok Nayak Hospital, described as abnormal lesions that emanate from Delhi, with chronic nasal obstruction, excessive any portion of the nasal mucosa or Para nasal sneezing, nasal discharge and frontal headache since sinuses. They are commonly located in the middle several months. Nasal obstruction was of gradual meatus and ethmoid sinus and are present in onset, non progressive, more on the left side than 1-4% of the population (1). -

Thesis FINAL PRINT
Preface This study was conducted at the Department of Chemistry, Biotechnology and Food Science (IKBM), Norwegian University of Life Sciences (UMB) during November 2009 to November 2010. My supervisors were Professor Dr Arne Tronsmo and PhD student Md. Hafizur Rahman, Department of IKBM, UMB. I express my sincere appreciation and gratitude to Professor Arne Tronsmo and Dr. Linda Gordon Hjeljord for their dynamic guidance throughout the period of the study, constant encouragement, constructive criticism and valuable suggestion during preparation of the thesis. I also want to thank Md. Hafizur Rahman for his guidance during my research work. Moreover I express my sincere gratitude to Grethe Kobro and Else Maria Aasen and all other staffs and workers at IKBM, UMB for their helpful co-operation to complete the research work in the laboratory. Last but not the least; I feel the heartiest indebtedness to Sabine and my family members for their patient inspirations, sacrifices and never ending encouragement. Ås, January 2011 Latifur Rahman Shovan i Abstract This thesis has been focused on methods to control diseases caused by Botrytis cinerea. B. cinerea causes grey mould disease of strawberry and chickpea, as well as many other plants. The fungal isolates used were isolated from chickpea leaf (Gazipur, Bangladesh) or obtained from the Norwegian culture collections of Bioforsk (Ås) and IKBM (UMB). Both morphological and molecular characterization helped to identify the fungal isolates as Botrytis cinerea (B. cinerea 101 and B. cinerea-BD), Trichoderma atroviride, T. asperellum Alternaria brassicicola, and Mucor piriformis. The identity of one fungal isolate, which was obtained from the culture collection of Bioforsk under the name Microdochium majus, could not be confirmed in this study. -
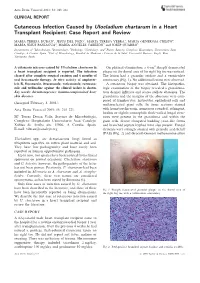
Cutaneous Infection Caused by Ulocladium Chartarum in a Heart Transplant Recipient: Case Report and Review
Acta Derm Venereol 2003; 83: 218–221 CLINICAL REPORT Cutaneous Infection Caused by Ulocladium chartarum in a Heart Transplant Recipient: Case Report and Review MARI´A TERESA DURA´ N1, JESU´ S DEL POZO2, MARI´A TERESA YEBRA3, MARI´A GENEROSA CRESPO4, MARI´A JESU´ S PANIAGUA4, MARI´A ANGELES CABEZO´ N5 and JOSEP GUARRO6 Departments of 1Microbiology, 2Dermatology, 3Pathology, 4Cardiology, and 5Plastic Surgery, Complexo Hospitalario Universitario Juan Canalejo, A Corun˜a, Spain, 6Unit of Microbiology, Facultat de Medicina i Cie`ncies de la Salut, Universitat Rovira i Virgili, Reus, Tarragona, Spain A cutaneous mycoses caused by Ulocladium chartarum in On physical examination, a 6-cm2 sharply demarcated a heart transplant recipient is reported. The infection plaque on the dorsal area of his right big toe was noticed. cleared after complete surgical excision and 6 months of The lesion had a granular surface and a vermiculate oral itraconazole therapy. In vitro activity of amphoter- consistency (Fig. 1). No additional lesions were observed. icin B, fluconazole, itraconazole, voriconazole, ravucona- A cutaneous biopsy was obtained. The histopatho- zole and terbinafine against the clinical isolate is shown. logic examination of the biopsy revealed a granuloma- Key words: dermatomycoses; immunocompromised host; tous dermal infiltrate and scarce stellate abscesses. The skin diseases. granuloma and the margins of the abscesses were com- posed of lymphocytes, histiocytes, epithelioid cells and (Accepted February 3, 2003.) multinucleated giant cells. In tissue sections stained Acta Derm Venereol 2003; 83: 218–221. with hematoxylin-eosin, numerous rounded, refringent, hyaline or slightly eosinophilic thick-walled fungal struc- Ma Teresa Dura´n Valle, Servicio de Microbiologı´a, tures were present in the granuloma and within the Complexo Hospitalario Universitario Juan Canalejo, giant cells. -

Novel Dematiaceous Hyphomycetes
STUDIES IN MYCOLOGY 50: 109–118. 2004. Novel dematiaceous hyphomycetes * Emory G. Simmons 717 Thornwood Road, Crawfordsville, Indiana 47933, U.S.A. *Correspondence: Emory G. Simmons, [email protected] Abstract: Novel taxa are described as Alternaria simsimi isolated from Sesamum indicum, distinguishing it from Alternaria sesami; Embellisia lolii from Lolium perenne; Nimbya perpunctulata from Alternanthera philoxeroides; Ulocladium arbores- cens from Pistacia vera; and Stemphylium symphyti from Symphytum × uplandicum. Taxonomic novelties: Alternaria simsimi E.G. Simmons sp. nov., Embellisia lolii E.G. Simmons & C.F. Hill sp. nov., Nimbya perpunctulata E.G. Simmons sp. nov., Stemphylium symphyti E.G. Simmons sp. nov., Ulocladium arborescens E.G. Simmons sp. nov. Key words: Alternaria, Embellisia, Nimbya, Ulocladium, Stemphylium, anamorphs, Sesamum, Lolium, Alternanthera, Pistacia, Symphytum. INTRODUCTION described and discussed on the basis of examination of colonies at 50× magnification and, at higher magnifi- It is a pleasure to be able to offer several new taxa in cations, of sporulation elements sampled at 5–7 d after honour of the centenary observances of the Centraal- inoculation. Each isolate or specimen discussed is bureau voor Schimmelcultures. These are dedicated to identified by a unique record/research number in the mycologist colleagues who were among my first format E.G.S. 00.000. associates during my earliest visits to CBS in Baarn, beginning in the late 1950s: Agathe van Beverwijk, Gerharda Bunschoten, G.A. de Vries, Amelia Stolk, TAXONOMIC PART Beatrice Schol-Schwarz, Maria Schipper, and Annie van der Plaats-Niterink. A novel taxon is presented for Alternaria: three species on sesame, two as patho- each of the five hyphomycetous genera that have been gens the major focus of my taxonomic work over the past Alternaria isolates from invaded tissues of Sesamum 50+ years. -

Monograph on Dematiaceous Fungi
Monograph On Dematiaceous fungi A guide for description of dematiaceous fungi fungi of medical importance, diseases caused by them, diagnosis and treatment By Mohamed Refai and Heidy Abo El-Yazid Department of Microbiology, Faculty of Veterinary Medicine, Cairo University 2014 1 Preface The first time I saw cultures of dematiaceous fungi was in the laboratory of Prof. Seeliger in Bonn, 1962, when I attended a practical course on moulds for one week. Then I handled myself several cultures of black fungi, as contaminants in Mycology Laboratory of Prof. Rieth, 1963-1964, in Hamburg. When I visited Prof. DE Varies in Baarn, 1963. I was fascinated by the tremendous number of moulds in the Centraalbureau voor Schimmelcultures, Baarn, Netherlands. On the other hand, I was proud, that El-Sheikh Mahgoub, a Colleague from Sundan, wrote an internationally well-known book on mycetoma. I have never seen cases of dematiaceous fungal infections in Egypt, therefore, I was very happy, when I saw the collection of mycetoma cases reported in Egypt by the eminent Egyptian Mycologist, Prof. Dr Mohamed Taha, Zagazig University. To all these prominent mycologists I dedicate this monograph. Prof. Dr. Mohamed Refai, 1.5.2014 Heinz Seeliger Heinz Rieth Gerard de Vries, El-Sheikh Mahgoub Mohamed Taha 2 Contents 1. Introduction 4 2. 30. The genus Rhinocladiella 83 2. Description of dematiaceous 6 2. 31. The genus Scedosporium 86 fungi 2. 1. The genus Alternaria 6 2. 32. The genus Scytalidium 89 2.2. The genus Aurobasidium 11 2.33. The genus Stachybotrys 91 2.3. The genus Bipolaris 16 2. -

PERSOONIAL R Eflections
Persoonia 23, 2009: 177–208 www.persoonia.org doi:10.3767/003158509X482951 PERSOONIAL R eflections Editorial: Celebrating 50 years of Fungal Biodiversity Research The year 2009 represents the 50th anniversary of Persoonia as the message that without fungi as basal link in the food chain, an international journal of mycology. Since 2008, Persoonia is there will be no biodiversity at all. a full-colour, Open Access journal, and from 2009 onwards, will May the Fungi be with you! also appear in PubMed, which we believe will give our authors even more exposure than that presently achieved via the two Editors-in-Chief: independent online websites, www.IngentaConnect.com, and Prof. dr PW Crous www.persoonia.org. The enclosed free poster depicts the 50 CBS Fungal Biodiversity Centre, Uppsalalaan 8, 3584 CT most beautiful fungi published throughout the year. We hope Utrecht, The Netherlands. that the poster acts as further encouragement for students and mycologists to describe and help protect our planet’s fungal Dr ME Noordeloos biodiversity. As 2010 is the international year of biodiversity, we National Herbarium of the Netherlands, Leiden University urge you to prominently display this poster, and help distribute branch, P.O. Box 9514, 2300 RA Leiden, The Netherlands. Book Reviews Mu«enko W, Majewski T, Ruszkiewicz- The Cryphonectriaceae include some Michalska M (eds). 2008. A preliminary of the most important tree pathogens checklist of micromycetes in Poland. in the world. Over the years I have Biodiversity of Poland, Vol. 9. Pp. personally helped collect populations 752; soft cover. Price 74 €. W. Szafer of some species in Africa and South Institute of Botany, Polish Academy America, and have witnessed the of Sciences, Lubicz, Kraków, Poland. -

Recurrence of Stachybotrys Chartarum During Mycological And
Recurrence of Stachybotrys chartarum during mycological and toxicological study of bioaerosols collected in a dairy cattle shed Caroline Lanier, Veronique Andre, Virginie Seguin, Natacha Heutte, Anne El Kaddoumi, Valérie Bouchart, Rachel Picquet, David Garon To cite this version: Caroline Lanier, Veronique Andre, Virginie Seguin, Natacha Heutte, Anne El Kaddoumi, et al.. Recur- rence of Stachybotrys chartarum during mycological and toxicological study of bioaerosols collected in a dairy cattle shed. Annals of Agricultural and Environmental Medicine (AAEM), Institute of Agricultural Medicine in Lublin, 2012, 19 (1), pp.61-67. hal-02186772 HAL Id: hal-02186772 https://hal-normandie-univ.archives-ouvertes.fr/hal-02186772 Submitted on 13 Nov 2019 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Distributed under a Creative Commons Attribution - NonCommercial| 4.0 International License Annals of Agricultural and Environmental Medicine 2012, Vol 19, No 1, 61-67 ORIGINAL ARTICLE www.aaem.pl Recurrence of Stachybotrys chartarum during mycological and toxicological study of bioaerosols collected in a dairy cattle shed Caroline Lanier1, Véronique André1, Virginie Séguin1, Natacha Heutte1, Anne El Kaddoumi1, Valérie Bouchart2, Rachel Picquet2, David Garon1 1 Université de Caen Basse Normandie, Caen, France 2 Laboratoire Départemental Frank Duncombe, Conseil Général du Calvados, Saint Contest, France Lanier C, André V, Séguin V, Heutte N, Kaddoumi A, Bouchart V, Picquet R, Garon D. -

Culture Inventory
For queries, contact the SFA leader: John Dunbar - [email protected] Fungal collection Putative ID Count Ascomycota Incertae sedis 4 Ascomycota Incertae sedis 3 Pseudogymnoascus 1 Basidiomycota Incertae sedis 1 Basidiomycota Incertae sedis 1 Capnodiales 29 Cladosporium 27 Mycosphaerella 1 Penidiella 1 Chaetothyriales 2 Exophiala 2 Coniochaetales 75 Coniochaeta 56 Lecythophora 19 Diaporthales 1 Prosthecium sp 1 Dothideales 16 Aureobasidium 16 Dothideomycetes incertae sedis 3 Dothideomycetes incertae sedis 3 Entylomatales 1 Entyloma 1 Eurotiales 393 Arthrinium 2 Aspergillus 172 Eladia 2 Emericella 5 Eurotiales 2 Neosartorya 1 Paecilomyces 13 Penicillium 176 Talaromyces 16 Thermomyces 4 Exobasidiomycetes incertae sedis 7 Tilletiopsis 7 Filobasidiales 53 Cryptococcus 53 Fungi incertae sedis 13 Fungi incertae sedis 12 Veroneae 1 Glomerellales 1 Glomerella 1 Helotiales 34 Geomyces 32 Helotiales 1 Phialocephala 1 Hypocreales 338 Acremonium 20 Bionectria 15 Cosmospora 1 Cylindrocarpon 2 Fusarium 45 Gibberella 1 Hypocrea 12 Ilyonectria 13 Lecanicillium 5 Myrothecium 9 Nectria 1 Pochonia 29 Purpureocillium 3 Sporothrix 1 Stachybotrys 3 Stanjemonium 2 Tolypocladium 1 Tolypocladium 2 Trichocladium 2 Trichoderma 171 Incertae sedis 20 Oidiodendron 20 Mortierellales 97 Massarineae 2 Mortierella 92 Mortierellales 3 Mortiererallales 2 Mortierella 2 Mucorales 109 Absidia 4 Backusella 1 Gongronella 1 Mucor 25 RhiZopus 13 Umbelopsis 60 Zygorhynchus 5 Myrmecridium 2 Myrmecridium 2 Onygenales 4 Auxarthron 3 Myceliophthora 1 Pezizales 2 PeZiZales 1 TerfeZia 1 -

Supplementary Table S1 18Jan 2021
Supplementary Table S1. Accurate scientific names of plant pathogenic fungi and secondary barcodes. Below is a list of the most important plant pathogenic fungi including Oomycetes with their accurate scientific names and synonyms. These scientific names include the results of the change to one scientific name for fungi. For additional information including plant hosts and localities worldwide as well as references consult the USDA-ARS U.S. National Fungus Collections (http://nt.ars- grin.gov/fungaldatabases/). Secondary barcodes, where available, are listed in superscript between round parentheses after generic names. The secondary barcodes listed here do not represent all known available loci for a given genus. Always consult recent literature for which primers and loci are required to resolve your species of interest. Also keep in mind that not all barcodes are available for all species of a genus and that not all species/genera listed below are known from sequence data. GENERA AND SPECIES NAME AND SYNONYMYS DISEASE SECONDARY BARCODES1 Kingdom Fungi Ascomycota Dothideomycetes Asterinales Asterinaceae Thyrinula(CHS-1, TEF1, TUB2) Thyrinula eucalypti (Cooke & Massee) H.J. Swart 1988 Target spot or corky spot of Eucalyptus Leptostromella eucalypti Cooke & Massee 1891 Thyrinula eucalyptina Petr. & Syd. 1924 Target spot or corky spot of Eucalyptus Lembosiopsis eucalyptina Petr. & Syd. 1924 Aulographum eucalypti Cooke & Massee 1889 Aulographina eucalypti (Cooke & Massee) Arx & E. Müll. 1960 Lembosiopsis australiensis Hansf. 1954 Botryosphaeriales Botryosphaeriaceae Botryosphaeria(TEF1, TUB2) Botryosphaeria dothidea (Moug.) Ces. & De Not. 1863 Canker, stem blight, dieback, fruit rot on Fusicoccum Sphaeria dothidea Moug. 1823 diverse hosts Fusicoccum aesculi Corda 1829 Phyllosticta divergens Sacc. 1891 Sphaeria coronillae Desm. -

Isolation and Characterization of the Mating-Type Locus of the Barley
855 Isolation and characterization of the mating-type locus of the barley pathogen Pyrenophora teres and frequencies of mating-type idiomorphs within and among fungal populations collected from barley landraces Domenico Rau, Frank J. Maier, Roberto Papa, Anthony H.D. Brown, Virgilio Balmas, Eva Saba, Wilhelm Schaefer, and Giovanna Attene Abstract: Pyrenophora teres f. sp. teres mating-type genes (MAT-1: 1190 bp; MAT-2: 1055 bp) have been identified. Their predicted proteins, measuring 379 and 333 amino acids, respectively, are similar to those of other Pleosporales, such as Pleospora sp., Cochliobolus sp., Alternaria alternata, Leptosphaeria maculans, and Phaeosphaeria nodorum. The structure of the MAT locus is discussed in comparison with those of other fungi. A mating-type PCR assay has also been developed; with this assay we have analyzed 150 isolates that were collected from 6 Sardinian barley land- race populations. Of these, 68 were P. t e re s f. sp. teres (net form; NF) and 82 were P. t e re s f. sp. maculata (spot form; SF). Within each mating type, the NF and SF amplification products were of the same length and were highly similar in sequence. The 2 mating types were present in both the NF and the SF populations at the field level, indicating that they have all maintained the potential for sexual reproduction. Despite the 2 forms being sympatric in 5 fields, no in- termediate isolates were detected with amplified fragment length polymorphism (AFLP) analysis. These results suggest that the 2 forms are genetically isolated under the field conditions. In all of the samples of P.