Biopython Tutorial and Cookbook
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Structuration Écologique Et Évolutive Des Symbioses Mycorhiziennes Des
Universit´ede La R´eunion { Ecole´ doctorale Sciences Technologie Sant´e{ Facult´edes Sciences et des Technologies Structuration ´ecologique et ´evolutive des symbioses mycorhiziennes des orchid´ees tropicales Th`ese de doctorat pr´esent´eeet soutenue publiquement le 19 novembre 2010 pour l'obtention du grade de Docteur de l'Universit´ede la R´eunion (sp´ecialit´eBiologie des Populations et Ecologie)´ par Florent Martos Composition du jury Pr´esidente: Mme Pascale Besse, Professeur `al'Universit´ede La R´eunion Rapporteurs : Mme Marie-Louise Cariou, Directrice de recherche au CNRS de Gif-sur-Yvette M. Raymond Tremblay, Professeur `al'Universit´ede Porto Rico Examinatrice : Mme Pascale Besse, Professeur `al'Universit´ede La R´eunion Directeurs : M. Thierry Pailler, Ma^ıtre de conf´erences HDR `al'Universit´ede La R´eunion M. Marc-Andr´e Selosse, Professeur `al'Universit´ede Montpellier Laboratoire d'Ecologie´ Marine Institut de Recherche pour le D´eveloppement 2 R´esum´e Les plantes n'exploitent pas seules les nutriments du sol, mais d´ependent de champignons avec lesquels elles forment des symbioses mycorhiziennes dans leurs racines. C'est en particulier vrai pour les 25 000 esp`ecesd'orchid´eesactuelles qui d´ependent toutes de champignons mycorhiziens pour accomplir leur cycle de vie. Elles produisent des graines microscopiques qui n'ont pas les ressources nutritives pour germer, mais qui d´ependent de la pr´esencede partenaires ad´equatspour nourrir l'embryon (h´et´erotrophie)jusqu’`al'apparition des feuilles (autotrophie). Les myco- rhiziens restent pr´esents dans les racines des adultes o`uils contribuent `ala nutrition, ce qui permet d'´etudierplus facilement la diversit´edes symbiotes `al'aide des ou- tils g´en´etiques.Conscients des biais des ´etudesen faveur des r´egionstemp´er´ees, nous avons ´etudi´ela diversit´edes mycorhiziens d'orchid´eestropicales `aLa R´eunion. -

Lepidosaphes Chinensis Chamberlin: Chinese Mussel Scale Hemiptera: Diaspididae Current Rating: Q Proposed Rating: A
CALIFORNIA DEPARTMENT OF cdfa FOOD & AGR I CULT URE ~ California Pest Rating Proposal Lepidosaphes chinensis Chamberlin: Chinese mussel scale Hemiptera: Diaspididae Current Rating: Q Proposed Rating: A Comment Period: 5/26/2021 – 7/10/2021 Initiating Event: Lepidosaphes chinensis is frequently intercepted in California on Dracaena plants from Florida, Ecuador, and Asia (China and Thailand). It was collected twice on orchids in Los Angeles County in 1934 and 1935 but it was since eradicated (Gill, 1997). It was also recently found in a nursery in Monterey County in 2011 (California Department of Food and Agriculture). It has not been rated. Therefore, a pest rating proposal is needed. History & Status: Background: The scale Lepidosaphes chinensis is reported to feed on plants in eight families: Arecaceae, Asparagaceae, Elaeagnaceae, Euphorbiaceae, Fabaceae, Magnoliaceae, Orchidaceae, and Pandanaceae (García Morales et al., 2016). Reported hosts Beaucarnea recurvata, Dracaena (including D. braunii, or lucky bamboo), Ficus, Sansevieriana trifasciata, and Yucca elephantipes (California Department of Food and Agriculture; Łabanowski, 2017; Suh and Bombay, 2015). Infestations are reported to cause chlorosis, necrosis, wilting, and black streaks, and heavy infestations cause the death of affected leaves (Malumphy et al., 2012; Stocks, 2014). No reports of economic damage were found, but it is apparent that the value of ornamental plants can be lowered, and this scale was deemed to have the potential to become a significant economic and ecological pest in Florida (Stocks, 2014). CALIFORNIA DEPARTMENT OF cdfa FOOD & AGR I CULT URE ~ Worldwide Distribution: Lepidosaphes chinensis is apparently native to eastern Asia, where it is reported from China, Hong Kong, Laos, Philippines, Singapore, Vietnam, Taiwan, and Thailand (Martin and Lau, 2011; García Morales et al., 2016; Suh and Bombay, 2015). -

July-August Bay Leaf
July-August 2005 The Bay Leaf California Native Plant Society • East Bay Chapter • Alameda & Contra Costa Counties www.ebcnps.org CALENDAR OF EVENTS Native Here p. 6 Saturday July 16, 10:00 am, field Trip to Cedar Fridays, July 1, 8, 15, 22, 29 and Aug 5, 12, 19, 26 Mountain Native Here Nursery open 9-noon Sunday July 24 at 2 pm, Pioneer Tree Trail, Samuel Saturdays, July 2, 9, 16, 23, 30 and Aug 6, 13, 20, Taylor State Park 27 nursery open 10-1 Saturday August 6 10:00 am, field trip onBlue Oak/ Tuesdays, July 5, 12, 19, 26 and Aug 2, 9, 16, 23, 30 Spengler Trail to Chaparral in Briones Regional Native Here seed collecting, 9 am Park Wednesday August 24 6:30 pm, keying session at Plant Sale Activities p. 6 Native Here Nursery Tuesdays, July 5, 12, 19, 26, 9 am to 2 pm, Merritt College, Oakland Board of Directors’ Meeting Saturday, August 13th, 10 am, Merritt College, Oak- Field Trips p. 5 land Saturday, July 9, 2005, Field Trip to Calaveras Big Trees State Park and Environs MOUNT DIABLO BUCKWHEAT REDISCOVERED Eriogonum truncatum has been dubbed the “Holy Grail I was working on the Mount Diablo project and target- of the East Bay” by Barbara Ertter. For such a delicate ing areas favorable to unusual plants. Further, a close plant, this name is somewhat deceptive. When Seth eye was to be kept for the diabolically elusive Mount Adams of Save Mount Diablo was brought to the site, Diablo Buckwheat that I believed was present in some he was unable to perceive the plant though it stood difficult to reach spot on the mountain. -

Special Status Vascular Plant Surveys and Habitat Modeling in Yosemite National Park, 2003–2004
National Park Service U.S. Department of the Interior Natural Resource Program Center Special Status Vascular Plant Surveys and Habitat Modeling in Yosemite National Park, 2003–2004 Natural Resource Technical Report NPS/SIEN/NRTR—2010/389 ON THE COVER USGS and NPS joint survey for Tompkins’ sedge (Carex tompkinsii), south side Merced River, El Portal, Mariposa County, California (upper left); Yosemite onion (Allium yosemitense) (upper right); Yosemite lewisia (Lewisia disepala) (lower left); habitat model for mountain lady’s slipper (Cypripedium montanum) in Yosemite National Park, California (lower right). Photographs by: Peggy E. Moore. Special Status Vascular Plant Surveys and Habitat Modeling in Yosemite National Park, 2003–2004 Natural Resource Technical Report NPS/SIEN/NRTR—2010/389 Peggy E. Moore, Alison E. L. Colwell, and Charlotte L. Coulter U.S. Geological Survey Western Ecological Research Center 5083 Foresta Road El Portal, California 95318 October 2010 U.S. Department of the Interior National Park Service Natural Resource Program Center Fort Collins, Colorado The National Park Service, Natural Resource Program Center publishes a range of reports that address natural resource topics of interest and applicability to a broad audience in the National Park Service and others in natural resource management, including scientists, conservation and environmental constituencies, and the public. The Natural Resource Technical Report Series is used to disseminate results of scientific studies in the physical, biological, and social sciences for both the advancement of science and the achievement of the National Park Service mission. The series provides contributors with a forum for displaying comprehensive data that are often deleted from journals because of page limitations. -

Ukiah Western Hills Open Land Acquisition & Limited Development
Ukiah Western Hills Open Land Acquisition & Limited Development Agreement Draft Initial Study & Mitigated Negative Declaration Attachments April 16, 2021 ATTACHMENT A Existing Site Photographs Existing access road Existing water tank site Existing "house site" on one of the proposed Development Parcels ATTACHMENT B Prepared For: Michelle Irace, Planning Manager Department of Community Development 300 Seminary Avenue, Ukiah, CA 95482 APNs: 001-040-83, 157- 070-01, 157-070-02, and 003-190-01 Prepared by Jacobszoon & Associates, Inc. Alicia Ives Ringstad Senior Wildlife Biologist [email protected] Date: March 11, 2021 Updated: April 8, 2021 Biological Assessment Report Table of Contents Section 1.0: Introduction .................................................................................................................................................................. 2 Section 2.0: Regulations and Descriptions ....................................................................................................................................... 2 2.1 Regulatory Setting ................................................................................................................................................................. 2 2.2 Natural Communities and Sensitive Natural Communities .................................................................................................... 3 2.3 Special-Status Species........................................................................................................................................................... -

Hunter Creek Nancy J
Hunter Creek Nancy J. Brian District Botanist, BLM Coos Bay District Office, North Bend, OR 97459 magine you can “apparate” (disappear from one place and reappear almost instantly in Ianother, like the wizards and witches in the Harry Potter series) to the middle of Hunter Creek Bog. As you slowly sank into the bog, you’d find yourself surrounded by California pitcherplant, pleated and Mendocino gentians, Labrador tea, western tofieldia, California bog asphodel, sedges, Columbia and Vollmer’s lilies, and Pacific reed grass. Outside of this watery jungle, you would see dry, bare, red rocky slopes and a scattering of Port Orford cedar and Jeffrey pine. Luckily, “muggles” (the non-magical community) don’t need to “apparate” to get to Hunter Creek Bog. We can turn off Highway 101, just south of Gold Beach in Curry County, and drive east about 10.5 miles to visit this public land botanical jewel managed by the Bureau of Land Management (BLM). ACEC Status There are two Areas of Critical Environmental Concern (ACEC) at Hunter Creek: Hunter Creek Bog ACEC and the North Fork Hunter Creek ACEC, which together total about 2,300 acres. In 1982 the Kalmiopsis Audubon Society and the Innominata Garden Club nominated Hunter Creek Bog (Section 13) and Hunter Springs Bog (Section 24) for ACEC status (Bowen and others 1982). In 1994, 570 acres in those sections and 1,730 acres of North Fork Hunter Creek were proposed as a separate ACEC. In 1995, both ACEC proposals were designated, including lands located in Sections 1, 2, 11, 12, 13, 14, and 24, Township 37 South, Range 14 West (BLM 1995). -

Download Flowering Plants of the Rough & Ready Creek Watershed List
FERNS & FERN ALLIES: DENNSTAEDTIACEAE BRACKEN FAMILY 1. Pteridium aquilinum var. pubescens Western Bracken Fern DRYOPTERIDACEAE WOOD FERN FAMILY Flowering Plants of the Rough & Ready Creek 2. Polystichum imbricans ssp. imbricans Imbricated or Narrow-leaved Sword Fern —Feb. 2015 revision of nomenclature based on the Watershed 3. Polystichum munitum Common Swordfern most current, third, Angiosperm Phylogeny Group (APG3) treatment of classification. Previously used names shown in EQUISETACEAE HORSETAIL FAMILY (parenthesizes). 4. Equisetum laevigatum Smooth Scouring Rush This plant list was originally a compilation of a variety of Rough and Ready area plant lists--compiled by Karen Phillips and Wendell Wood. Lists include POLYPODIACEAE POLYPODY FAMILY species contained in Barbara Ullian's "Preliminary Flora - Rough & Ready Creek" 5. Polypodium glycyrrhiza Licorice Fern (1994) which includes species recorded by Mary Paetzel and Mike Anderson. Other Rough and Ready lists were previously contributed by Veva Stansell, PTERIDACEAE BRAKE FAMILY Robin Taylor-Davenport and Jill Pade. Additionally, species were added as 6. Adiantum aleuticum (pedatum) Five-fingered Fern or Maidenhair Fern contained on The Nature Conservancy preserve’s 2007 list, and species identified in 2008-2009 for the Medford Dist. BLM’s Rough and Ready 7. Aspidotis densa Indian’s Dream ACEC by botanical contractors: Scot Loring, Josh Paque and Pete Kaplowe. 8. Pentagramma triangularis ssp. triangularis Goldback Fern Finally, Wendell Wood has added additional species he has personally found and identified in the Rough and Ready watershed prior to 2015. GYMNOSPERMS: CUPRESSACEAE CYPRESS FAMILY A few species on previous lists are shown separately after the end of this list, if they are not shown as occurring in Josephine Co. -

NPSO Annual Meeting July 13-15, 2012
NPSO Annual Meeting July 13-15, 2012 The Siskiyou Chapter invites you to enjoy the botanical wonders of southwestern Oregon at the NPSO Annual Meeting to be staged out of the Siskiyou Field Institute's Deer Creek Center. Formerly a ranch along Deer Creek on the north side of Eight Dollar Mountain, this environmental education center is only a few miles west of Selma in the Illinois River Valley, Josephine County. In all directions, the diverse flora of this region awaits your exploration on foot or by vehicle. Wade across Deer Creek and stand in an extensive series of fens at the foot of Eight Dollar Mountain, dominated by forbs and graminoids, including rare endemic sedges. The most conspicuous inhabitant is California pitcher plant (Darlingtonia californica); its two-foot- tall cylindrical leaves contain a watery soup of dead and decaying insects. Other spectacular plants associated with these fens are Waldo gentian (Gentiana setigera), Vollmer's lily (Lilium pardalinum ssp. vollmeri), large flowered bog lily, (Hastingsia bracteosa), yellow bog asphodel (Narthecium califoricum), western azalea (Rhododendron occidentale), California coneflower (Rudbeckia califonica) and California lady slipper (Cypripedium californicum). Southwestern Oregon has attracted botanists since the first European expeditions explored the Western region. John Jeffrey probably passed through here in 1851 on his way to Crescent City, California; several species bear his name, e.g., Pinus jeffreyi, Dodecatheon jeffreyi, Penstemon jeffreyanus. Thomas Jefferson Howell, a self-trained botanist and author of the first regional flora, made three collecting trips in the Illinois Valley in the 1870s, arriving as soon as the railroad reached Grants Pass. -
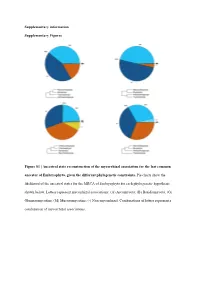
Ancestral State Reconstruction of the Mycorrhizal Association for the Last Common Ancestor of Embryophyta, Given the Different Phylogenetic Constraints
Supplementary information Supplementary Figures Figure S1 | Ancestral state reconstruction of the mycorrhizal association for the last common ancestor of Embryophyta, given the different phylogenetic constraints. Pie charts show the likelihood of the ancestral states for the MRCA of Embryophyta for each phylogenetic hypothesis shown below. Letters represent mycorrhizal associations: (A) Ascomycota; (B) Basidiomycota; (G) Glomeromycotina; (M) Mucoromycotina; (-) Non-mycorrhizal. Combinations of letters represent a combination of mycorrhizal associations. Austrocedrus chilensis Chamaecyparis obtusa Sequoiadendron giganteum Prumnopitys taxifolia Prumnopitys Prumnopitys montana Prumnopitys Prumnopitys ferruginea Prumnopitys Araucaria angustifolia Araucaria Dacrycarpus dacrydioides Dacrycarpus Taxus baccata Podocarpus oleifolius Podocarpus Afrocarpus falcatus Afrocarpus Ephedra fragilis Nymphaea alba Nymphaea Gnetum gnemon Abies alba Abies balsamea Austrobaileya scandens Austrobaileya Abies nordmanniana Thalictrum minus Thalictrum Abies homolepis Caltha palustris Caltha Abies magnifica ia repens Ranunculus Abies religiosa Ranunculus montanus Ranunculus Clematis vitalba Clematis Keteleeria davidiana Anemone patens Anemone Tsuga canadensis Vitis vinifera Vitis Tsuga mertensiana Saxifraga oppositifolia Saxifraga Larix decidua Hypericum maculatum Hypericum Larix gmelinii Phyllanthus calycinus Phyllanthus Larix kaempferi Hieronyma oblonga Hieronyma Pseudotsuga menziesii Salix reinii Salix Picea abies Salix polaris Salix Picea crassifolia Salix herbacea -

Phylogenetics, Genome Size Evolution and Population Ge- Netics of Slipper Orchids in the Subfamily Cypripedioideae (Orchidaceae)
ORBIT - Online Repository of Birkbeck Institutional Theses Enabling Open Access to Birkbecks Research Degree output Phylogenetics, genome size evolution and population ge- netics of slipper orchids in the subfamily cypripedioideae (orchidaceae) http://bbktheses.da.ulcc.ac.uk/88/ Version: Full Version Citation: Chochai, Araya (2014) Phylogenetics, genome size evolution and pop- ulation genetics of slipper orchids in the subfamily cypripedioideae (orchidaceae). PhD thesis, Birkbeck, University of London. c 2014 The Author(s) All material available through ORBIT is protected by intellectual property law, including copyright law. Any use made of the contents should comply with the relevant law. Deposit guide Contact: email Phylogenetics, genome size evolution and population genetics of slipper orchids in the subfamily Cypripedioideae (Orchidaceae) Thesis submitted by Araya Chochai For the degree of Doctor of Philosophy School of Science Birkbeck, University of London and Genetic Section, Jodrell Laboratory Royal Botanic Gardens, Kew November, 2013 Declaration I hereby confirm that this thesis is my own work and the material from other sources used in this work has been appropriately and fully acknowledged. Araya Chochai London, November 2013 2 Abstract Slipper orchids (subfamily Cypripedioideae) comprise five genera; Paphiopedilum, Cypripedium, Phragmipedium, Selenipedium, and Mexipedium. Phylogenetic relationships of the genus Paphiopedilum, were studied using nuclear ribosomal ITS and plastid sequence data. The results confirm that Paphiopedilum is monophyletic and support the division of the genus into three subgenera Parvisepalum, Brachypetalum and Paphiopedilum. Four sections of subgenus Paphiopedilum (Pardalopetalum, Cochlopetalum, Paphiopedilum and Barbata) are recovered with strong support for monophyly, concurring with a recent infrageneric treatment. Section Coryopedilum is also recovered with low bootstrap but high posterior probability values. -

Rare and Endangered Plants and Timber Harvest Plans In
RARE AND ENDANGERED PLANT GUIDE for TIMBER HARVEST PLANS in COASTAL MENDOCINO COUNTY (Office Manual) By Gordon E. McBride, PhD. Edited by Tamara Camper, Shayne Green, Darcie Mahoney and Geri Hulse-Stephens TABLE OF CONTENTS INTRODUCTION………………………………………………………………………...3 THE FLORA OF CALIFORNIA AND MENDOCINO COUNTY………………………4 AUTHORITIES AND AGENCIES………………………………………………..........4-9 DATABASES…………………………………………………………………………....10 SCOPING………………………………………………………………………….…….11 FIELD SEARCH PARAMETERS………………………………………………………11 PLANT IDENTIFICATION REFERENCES AND KEYS………………………….12-14 HERBARIA……………………………………………………………………………...14 I FOUND IT – NOW WHAT????.....................................................................................15 THE STEWARDSHIP ETHIC………………………………………….……………….16 REFERENCES……………………………………………………………………….......17 Revised March 15, 2017 INTRODUCTION The purpose of this manual is to provide guidance for Registered Professional Foresters (RPFs) faced with discovery, identification, and protection of rare or endangered plants. When preparing a Timber Harvest Plan (THP), the RPF must address these botanical resources per the criteria of CalFire, California Department of Fish and Wildlife (DFW) and other public and private groups. This manual will review: • Authorities that deal with rare or endangered plants • Databases used to generate information on these plants • Field search parameters • Survey reports • Plant identification references • Herbarium use Accompanying the office manual is a field guide (download a free copy) that provides descriptions, -

CITES and Slipper Orchids
CITES and Slipper Orchids An introduction to slipper orchids covered by the Convention on International Trade in Endangered Species Written by H. Noel McGough, David L. Roberts, Chris Brodie and Jenny Kowalczyk Royal Botanic Gardens, Kew United Kingdom The Board of Trustees, Royal Botanic Gardens, Kew 2006 © The Board of Trustees of the Royal Botanic Gardens, Kew 2006 All rights reserved. No part of this publication may be reproduced, stored in a retrieval system, or transmitted, in any form, or by any means, electronic, mechanical, photocopying, recording or otherwise, without written permission of the publisher unless in accordance with the provisions of the Copyright Designs and Patents Act 1988. First published in 2006 by Royal Botanic Gardens, Kew Richmond, Surrey, TW9 3AB, UK www.kew.org ISBN 1-84246-128-1 For information or to purchase Kew titles please visit www.kewbooks.com or email [email protected] Cover image: © Royal Botanic Gardens, Kew CONTENTS Introduction ..................................................................................................... i Acknowledgements ........................................................................................ ii How to Use this Presentation Pack ............................................................... iii References and Resources ........................................................................ iv-ix Slide Index ................................................................................................. x-xi Slides and speaker’s notes .......................................................................