Await Remainder of Exome Sequence Data
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Rare Modifier Variants Alter the Severity of Cardiovascular Disease in Pseudoxanthoma Elasticum
fcell-09-612581 June 8, 2021 Time: 11:34 # 1 BRIEF RESEARCH REPORT published: 08 June 2021 doi: 10.3389/fcell.2021.612581 Rare Modifier Variants Alter the Severity of Cardiovascular Disease in Pseudoxanthoma Elasticum: Identification of Novel Candidate Modifier Genes and Disease Pathways Through Mixture of Effects Analysis Eva Y. G. De Vilder1,2,3, Ludovic Martin4, Georges Lefthériotis5, Paul Coucke1,6, Filip Van Nieuwerburgh7 and Olivier M. Vanakker1,6* Edited by: 1 2 Svetlana Komarova, Center for Medical Genetics, Ghent University Hospital, Ghent, Belgium, The Research Foundation – Flanders, Ghent, 3 4 McGill University, Canada Belgium, Department of Ophthalmology, Ghent University Hospital, Ghent, Belgium, Department of Dermatology, Angers University Hospital, Angers, France, 5 Department of Vascular Physiology and Sports Medicine, Angers University, Angers, Reviewed by: France, 6 Department of Biomolecular Medicine, Ghent University, Ghent, Belgium, 7 Department of Pharmaceutics, Carolina Beraldo Meloto, Laboratory of Pharmaceutical Biotechnology, Ghent University, Ghent, Belgium McGill University, Canada Koen van de Wetering, Thomas Jefferson University, Introduction: Pseudoxanthoma elasticum (PXE), an ectopic mineralization disorder United States caused by pathogenic ABCC6 variants, is characterized by skin, ocular and Jessica Bertrand, University Hospital Magdeburg, cardiovascular (CV) symptoms. Due to striking phenotypic variability without genotype- Germany phenotype correlations, modifier genes are thought to play a role in disease variability. *Correspondence: In this study, we evaluated the collective modifying effect of rare variants on the Olivier M. Vanakker [email protected] cardiovascular phenotype of PXE. Materials and Methods: Mixed effects of rare variants were assessed by Whole Exome Specialty section: This article was submitted to Sequencing in 11 PXE patients with an extreme CV phenotype (mild/severe). -

Genetic Variation Across the Human Olfactory Receptor Repertoire Alters Odor Perception
bioRxiv preprint doi: https://doi.org/10.1101/212431; this version posted November 1, 2017. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. Genetic variation across the human olfactory receptor repertoire alters odor perception Casey Trimmer1,*, Andreas Keller2, Nicolle R. Murphy1, Lindsey L. Snyder1, Jason R. Willer3, Maira Nagai4,5, Nicholas Katsanis3, Leslie B. Vosshall2,6,7, Hiroaki Matsunami4,8, and Joel D. Mainland1,9 1Monell Chemical Senses Center, Philadelphia, Pennsylvania, USA 2Laboratory of Neurogenetics and Behavior, The Rockefeller University, New York, New York, USA 3Center for Human Disease Modeling, Duke University Medical Center, Durham, North Carolina, USA 4Department of Molecular Genetics and Microbiology, Duke University Medical Center, Durham, North Carolina, USA 5Department of Biochemistry, University of Sao Paulo, Sao Paulo, Brazil 6Howard Hughes Medical Institute, New York, New York, USA 7Kavli Neural Systems Institute, New York, New York, USA 8Department of Neurobiology and Duke Institute for Brain Sciences, Duke University Medical Center, Durham, North Carolina, USA 9Department of Neuroscience, University of Pennsylvania School of Medicine, Philadelphia, Pennsylvania, USA *[email protected] ABSTRACT The human olfactory receptor repertoire is characterized by an abundance of genetic variation that affects receptor response, but the perceptual effects of this variation are unclear. To address this issue, we sequenced the OR repertoire in 332 individuals and examined the relationship between genetic variation and 276 olfactory phenotypes, including the perceived intensity and pleasantness of 68 odorants at two concentrations, detection thresholds of three odorants, and general olfactory acuity. -

Qt4vh1p2c4 Nosplash E372185
Copyright 2014 by Janine Micheli-Jazdzewski ii Dedication I would like to dedicate this thesis to Rock, who is not with us anymore, TR, General Jack D. Ripper, and Page. Thank you for sitting with me while I worked for countless hours over the years. iii Acknowledgements I would like to express my special appreciation and thanks to my advisor Dr. Deanna Kroetz, you have been a superb mentor for me. I would like to thank you for encouraging my research and for helping me to grow as a research scientist. Your advice on both research, as well as on my career have been priceless. I would also like to thank my committee members, Dr. Laura Bull, Dr. Steve Hamilton and Dr. John Witte for guiding my research and expanding my knowledge on statistics, genetics and clinical phenotypes. I also want to thank past and present members of my laboratory for their support and help over the years, especially Dr. Mike Baldwin, Dr. Sveta Markova, Dr. Ying Mei Liu and Dr. Leslie Chinn. Thanks are also due to my many collaborators that made this research possible including: Dr. Eric Jorgenson, Dr. David Bangsberg, Dr. Taisei Mushiroda, Dr. Michiaki Kubo, Dr. Yusuke Nakamura, Dr. Jeffrey Martin, Joel Mefford, Dr. Sarah Shutgarts, Dr. Sulggi Lee and Dr. Sook Wah Yee. A special thank you to the RIKEN Center for Genomic Medicine that generously performed the genome-wide genotyping for these projects. Thanks to Dr. Steve Chamow, Dr. Bill Werner, Dr. Montse Carrasco, and Dr. Teresa Chen who started me on the path to becoming a scientist. -

Identification of 42 Genes Linked to Stage II Colorectal Cancer Metastatic Relapse
Int. J. Mol. Sci. 2016, 17, 598; doi:10.3390/ijms17040598 S1 of S16 Supplementary Materials: Identification of 42 Genes Linked to Stage II Colorectal Cancer Metastatic Relapse Rabeah A. Al-Temaimi, Tuan Zea Tan, Makia J. Marafie, Jean Paul Thiery, Philip Quirke and Fahd Al-Mulla Figure S1. Cont. Int. J. Mol. Sci. 2016, 17, 598; doi:10.3390/ijms17040598 S2 of S16 Figure S1. Mean expression levels of fourteen genes of significant association with CRC DFS and OS that are differentially expressed in normal colon compared to CRC tissues. Each dot represents a sample. Table S1. Copy number aberrations associated with poor disease-free survival and metastasis in early stage II CRC as predicted by STAC and SPPS combined methodologies with resident gene symbols. CN stands for copy number, whereas CNV is copy number variation. Region Cytoband % of CNV Count of Region Event Gene Symbols Length Location Overlap Genes chr1:113,025,076–113,199,133 174,057 p13.2 CN Loss 0.0 2 AKR7A2P1, SLC16A1 chr1:141,465,960–141,822,265 356,305 q12–q21.1 CN Gain 95.9 1 SRGAP2B MIR5087, LOC10013000 0, FLJ39739, LOC10028679 3, PPIAL4G, PPIAL4A, NBPF14, chr1:144,911,564–146,242,907 1,331,343 q21.1 CN Gain 99.6 16 NBPF15, NBPF16, PPIAL4E, NBPF16, PPIAL4D, PPIAL4F, LOC645166, LOC388692, FCGR1C chr1:177,209,428–177,226,812 17,384 q25.3 CN Gain 0.0 0 chr1:197,652,888–197,676,831 23,943 q32.1 CN Gain 0.0 1 KIF21B chr1:201,015,278–201,033,308 18,030 q32.1 CN Gain 0.0 1 PLEKHA6 chr1:201,289,154–201,298,247 9093 q32.1 CN Gain 0.0 0 chr1:216,820,186–217,043,421 223,235 q41 CN -

The Hypothalamus As a Hub for SARS-Cov-2 Brain Infection and Pathogenesis
bioRxiv preprint doi: https://doi.org/10.1101/2020.06.08.139329; this version posted June 19, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. The hypothalamus as a hub for SARS-CoV-2 brain infection and pathogenesis Sreekala Nampoothiri1,2#, Florent Sauve1,2#, Gaëtan Ternier1,2ƒ, Daniela Fernandois1,2 ƒ, Caio Coelho1,2, Monica ImBernon1,2, Eleonora Deligia1,2, Romain PerBet1, Vincent Florent1,2,3, Marc Baroncini1,2, Florence Pasquier1,4, François Trottein5, Claude-Alain Maurage1,2, Virginie Mattot1,2‡, Paolo GiacoBini1,2‡, S. Rasika1,2‡*, Vincent Prevot1,2‡* 1 Univ. Lille, Inserm, CHU Lille, Lille Neuroscience & Cognition, DistAlz, UMR-S 1172, Lille, France 2 LaBoratorY of Development and PlasticitY of the Neuroendocrine Brain, FHU 1000 daYs for health, EGID, School of Medicine, Lille, France 3 Nutrition, Arras General Hospital, Arras, France 4 Centre mémoire ressources et recherche, CHU Lille, LiCEND, Lille, France 5 Univ. Lille, CNRS, INSERM, CHU Lille, Institut Pasteur de Lille, U1019 - UMR 8204 - CIIL - Center for Infection and ImmunitY of Lille (CIIL), Lille, France. # and ƒ These authors contriButed equallY to this work. ‡ These authors directed this work *Correspondence to: [email protected] and [email protected] Short title: Covid-19: the hypothalamic hypothesis 1 bioRxiv preprint doi: https://doi.org/10.1101/2020.06.08.139329; this version posted June 19, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. -

Lupus Nephritis Supp Table 5
Supplementary Table 5 : Transcripts and DAVID pathways correlating with the expression of CD4 in lupus kidney biopsies Positive correlation Negative correlation Transcripts Pathways Transcripts Pathways Identifier Gene Symbol Correlation coefficient with CD4 Annotation Cluster 1 Enrichment Score: 26.47 Count P_Value Benjamini Identifier Gene Symbol Correlation coefficient with CD4 Annotation Cluster 1 Enrichment Score: 3.16 Count P_Value Benjamini ILMN_1727284 CD4 1 GOTERM_BP_FAT translational elongation 74 2.50E-42 1.00E-38 ILMN_1681389 C2H2 zinc finger protein-0.40001984 INTERPRO Ubiquitin-conjugating enzyme/RWD-like 17 2.00E-05 4.20E-02 ILMN_1772218 HLA-DPA1 0.934229063 SP_PIR_KEYWORDS ribosome 60 2.00E-41 4.60E-39 ILMN_1768954 RIBC1 -0.400186083 SMART UBCc 14 1.00E-04 3.50E-02 ILMN_1778977 TYROBP 0.933302249 KEGG_PATHWAY Ribosome 65 3.80E-35 6.60E-33 ILMN_1699190 SORCS1 -0.400223681 SP_PIR_KEYWORDS ubl conjugation pathway 81 1.30E-04 2.30E-02 ILMN_1689655 HLA-DRA 0.915891173 SP_PIR_KEYWORDS protein biosynthesis 91 4.10E-34 7.20E-32 ILMN_3249088 LOC93432 -0.400285215 GOTERM_MF_FAT small conjugating protein ligase activity 35 1.40E-04 4.40E-02 ILMN_3228688 HLA-DRB1 0.906190291 SP_PIR_KEYWORDS ribonucleoprotein 114 4.80E-34 6.70E-32 ILMN_1680436 CSH2 -0.400299744 SP_PIR_KEYWORDS ligase 54 1.50E-04 2.00E-02 ILMN_2157441 HLA-DRA 0.902996561 GOTERM_CC_FAT cytosolic ribosome 59 3.20E-33 2.30E-30 ILMN_1722755 KRTAP6-2 -0.400334007 GOTERM_MF_FAT acid-amino acid ligase activity 40 1.60E-04 4.00E-02 ILMN_2066066 HLA-DRB6 0.901531942 SP_PIR_KEYWORDS -

Us 2018 / 0305689 A1
US 20180305689A1 ( 19 ) United States (12 ) Patent Application Publication ( 10) Pub . No. : US 2018 /0305689 A1 Sætrom et al. ( 43 ) Pub . Date: Oct. 25 , 2018 ( 54 ) SARNA COMPOSITIONS AND METHODS OF plication No . 62 /150 , 895 , filed on Apr. 22 , 2015 , USE provisional application No . 62/ 150 ,904 , filed on Apr. 22 , 2015 , provisional application No. 62 / 150 , 908 , (71 ) Applicant: MINA THERAPEUTICS LIMITED , filed on Apr. 22 , 2015 , provisional application No. LONDON (GB ) 62 / 150 , 900 , filed on Apr. 22 , 2015 . (72 ) Inventors : Pål Sætrom , Trondheim (NO ) ; Endre Publication Classification Bakken Stovner , Trondheim (NO ) (51 ) Int . CI. C12N 15 / 113 (2006 .01 ) (21 ) Appl. No. : 15 /568 , 046 (52 ) U . S . CI. (22 ) PCT Filed : Apr. 21 , 2016 CPC .. .. .. C12N 15 / 113 ( 2013 .01 ) ; C12N 2310 / 34 ( 2013. 01 ) ; C12N 2310 /14 (2013 . 01 ) ; C12N ( 86 ) PCT No .: PCT/ GB2016 /051116 2310 / 11 (2013 .01 ) $ 371 ( c ) ( 1 ) , ( 2 ) Date : Oct . 20 , 2017 (57 ) ABSTRACT The invention relates to oligonucleotides , e . g . , saRNAS Related U . S . Application Data useful in upregulating the expression of a target gene and (60 ) Provisional application No . 62 / 150 ,892 , filed on Apr. therapeutic compositions comprising such oligonucleotides . 22 , 2015 , provisional application No . 62 / 150 ,893 , Methods of using the oligonucleotides and the therapeutic filed on Apr. 22 , 2015 , provisional application No . compositions are also provided . 62 / 150 ,897 , filed on Apr. 22 , 2015 , provisional ap Specification includes a Sequence Listing . SARNA sense strand (Fessenger 3 ' SARNA antisense strand (Guide ) Mathew, Si Target antisense RNA transcript, e . g . NAT Target Coding strand Gene Transcription start site ( T55 ) TY{ { ? ? Targeted Target transcript , e . -
Explorations in Olfactory Receptor Structure and Function by Jianghai
Explorations in Olfactory Receptor Structure and Function by Jianghai Ho Department of Neurobiology Duke University Date:_______________________ Approved: ___________________________ Hiroaki Matsunami, Supervisor ___________________________ Jorg Grandl, Chair ___________________________ Marc Caron ___________________________ Sid Simon ___________________________ [Committee Member Name] Dissertation submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy in the Department of Neurobiology in the Graduate School of Duke University 2014 ABSTRACT Explorations in Olfactory Receptor Structure and Function by Jianghai Ho Department of Neurobiology Duke University Date:_______________________ Approved: ___________________________ Hiroaki Matsunami, Supervisor ___________________________ Jorg Grandl, Chair ___________________________ Marc Caron ___________________________ Sid Simon ___________________________ [Committee Member Name] An abstract of a dissertation submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy in the Department of Neurobiology in the Graduate School of Duke University 2014 Copyright by Jianghai Ho 2014 Abstract Olfaction is one of the most primitive of our senses, and the olfactory receptors that mediate this very important chemical sense comprise the largest family of genes in the mammalian genome. It is therefore surprising that we understand so little of how olfactory receptors work. In particular we have a poor idea of what chemicals are detected by most of the olfactory receptors in the genome, and for those receptors which we have paired with ligands, we know relatively little about how the structure of these ligands can either activate or inhibit the activation of these receptors. Furthermore the large repertoire of olfactory receptors, which belong to the G protein coupled receptor (GPCR) superfamily, can serve as a model to contribute to our broader understanding of GPCR-ligand binding, especially since GPCRs are important pharmaceutical targets. -

Investigating the Effects of Copy Number Variants on Reading and Language Performance Alessandro Gialluisi1,2, Alessia Visconti3, Erik G
Gialluisi et al. Journal of Neurodevelopmental Disorders (2016) 8:17 DOI 10.1186/s11689-016-9147-8 RESEARCH Open Access Investigating the effects of copy number variants on reading and language performance Alessandro Gialluisi1,2, Alessia Visconti3, Erik G. Willcutt4,5, Shelley D. Smith6, Bruce F. Pennington7, Mario Falchi3, John C. DeFries4,5, Richard K. Olson4,5, Clyde Francks1,8* and Simon E. Fisher1,8* Abstract Background: Reading and language skills have overlapping genetic bases, most of which are still unknown. Part of the missing heritability may be caused by copy number variants (CNVs). Methods: In a dataset of children recruited for a history of reading disability (RD, also known as dyslexia) or attention deficit hyperactivity disorder (ADHD) and their siblings, we investigated the effects of CNVs on reading and language performance. First, we called CNVs with PennCNV using signal intensity data from Illumina OmniExpress arrays (~723,000 probes). Then, we computed the correlation between measures of CNV genomic burden and the first principal component (PC) score derived from several continuous reading and language traits, both before and after adjustment for performance IQ. Finally, we screened the genome, probe-by-probe, for association with the PC scores, through two complementary analyses: we tested a binary CNV state assigned for the location of each probe (i.e., CNV+ or CNV−), and we analyzed continuous probe intensity data using FamCNV. Results: No significant correlation was found between measures of CNV burden and PC scores, and no genome-wide significant associations were detected in probe-by-probe screening. Nominally significant associations were detected (p~10−2–10−3)withinCNTN4 (contactin 4) and CTNNA3 (catenin alpha 3). -

Online Supporting Information S2: Proteins in Each Negative Pathway
Online Supporting Information S2: Proteins in each negative pathway Index Proteins ADO,ACTA1,DEGS2,EPHA3,EPHB4,EPHX2,EPOR,EREG,FTH1,GAD1,HTR6, IGF1R,KIR2DL4,NCR3,NME7,NOTCH1,OR10S1,OR2T33,OR56B4,OR7A10, Negative_1 OR8G1,PDGFC,PLCZ1,PROC,PRPS2,PTAFR,SGPP2,STMN1,VDAC3,ATP6V0 A1,MAPKAPK2 DCC,IDS,VTN,ACTN2,AKR1B10,CACNA1A,CHIA,DAAM2,FUT5,GCLM,GNAZ Negative_2 ,ITPA,NEU4,NTF3,OR10A3,PAPSS1,PARD3,PLOD1,RGS3,SCLY,SHC1,TN FRSF4,TP53 Negative_3 DAO,CACNA1D,HMGCS2,LAMB4,OR56A3,PRKCQ,SLC25A5 IL5,LHB,PGD,ADCY3,ALDH1A3,ATP13A2,BUB3,CD244,CYFIP2,EPHX2,F CER1G,FGD1,FGF4,FZD9,HSD17B7,IL6R,ITGAV,LEFTY1,LIPG,MAN1C1, Negative_4 MPDZ,PGM1,PGM3,PIGM,PLD1,PPP3CC,TBXAS1,TKTL2,TPH2,YWHAQ,PPP 1R12A HK2,MOS,TKT,TNN,B3GALT4,B3GAT3,CASP7,CDH1,CYFIP1,EFNA5,EXTL 1,FCGR3B,FGF20,GSTA5,GUK1,HSD3B7,ITGB4,MCM6,MYH3,NOD1,OR10H Negative_5 1,OR1C1,OR1E1,OR4C11,OR56A3,PPA1,PRKAA1,PRKAB2,RDH5,SLC27A1 ,SLC2A4,SMPD2,STK36,THBS1,SERPINC1 TNR,ATP5A1,CNGB1,CX3CL1,DEGS1,DNMT3B,EFNB2,FMO2,GUCY1B3,JAG Negative_6 2,LARS2,NUMB,PCCB,PGAM1,PLA2G1B,PLOD2,PRDX6,PRPS1,RFXANK FER,MVD,PAH,ACTC1,ADCY4,ADCY8,CBR3,CLDN16,CPT1A,DDOST,DDX56 ,DKK1,EFNB1,EPHA8,FCGR3A,GLS2,GSTM1,GZMB,HADHA,IL13RA2,KIR2 Negative_7 DS4,KLRK1,LAMB4,LGMN,MAGI1,NUDT2,OR13A1,OR1I1,OR4D11,OR4X2, OR6K2,OR8B4,OXCT1,PIK3R4,PPM1A,PRKAG3,SELP,SPHK2,SUCLG1,TAS 1R2,TAS1R3,THY1,TUBA1C,ZIC2,AASDHPPT,SERPIND1 MTR,ACAT2,ADCY2,ATP5D,BMPR1A,CACNA1E,CD38,CYP2A7,DDIT4,EXTL Negative_8 1,FCER1G,FGD3,FZD5,ITGAM,MAPK8,NR4A1,OR10V1,OR4F17,OR52D1,O R8J3,PLD1,PPA1,PSEN2,SKP1,TACR3,VNN1,CTNNBIP1 APAF1,APOA1,CARD11,CCDC6,CSF3R,CYP4F2,DAPK1,FLOT1,GSTM1,IL2 -

Jimmunol.1901059.Full.Pdf
Molecular Drivers of Lymphocyte Organization in Vertebrate Mucosal Surfaces: Revisiting the TNF Superfamily Hypothesis This information is current as of October 1, 2021. Ryan D. Heimroth, Elisa Casadei and Irene Salinas J Immunol published online 1 April 2020 http://www.jimmunol.org/content/early/2020/03/31/jimmun ol.1901059 Downloaded from Supplementary http://www.jimmunol.org/content/suppl/2020/03/31/jimmunol.190105 Material 9.DCSupplemental http://www.jimmunol.org/ Why The JI? Submit online. • Rapid Reviews! 30 days* from submission to initial decision • No Triage! Every submission reviewed by practicing scientists • Fast Publication! 4 weeks from acceptance to publication by guest on October 1, 2021 *average Subscription Information about subscribing to The Journal of Immunology is online at: http://jimmunol.org/subscription Permissions Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Email Alerts Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts The Journal of Immunology is published twice each month by The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2020 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. Published April 1, 2020, doi:10.4049/jimmunol.1901059 The Journal of Immunology Molecular Drivers of Lymphocyte Organization in Vertebrate Mucosal Surfaces: Revisiting the TNF Superfamily Hypothesis Ryan D. Heimroth, Elisa Casadei, and Irene Salinas The adaptive immune system of all jawed vertebrates relies on the presence of B and T cell lymphocytes that aggregate in specific body sites to form primary and secondary lymphoid structures. -
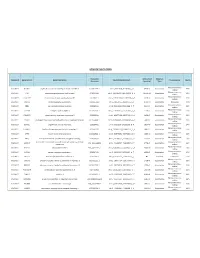
Somatic Mutations
SOMATIC MUTATIONS Transcript Amino Acid Mutation Sample ID Gene Symbol Gene Description Nucleotide (genomic) Consequence Mut % Accession (protein) Type Nonsynonymous PGDX11T ACSBG2 acyl-CoA synthetase bubblegum family member 2 CCDS12159.1 chr19_6141628_6141628_C_A 654A>D Substitution 46% coding Nonsynonymous PGDX11T ATR ataxia telangiectasia and Rad3 related CCDS3124.1 chr3_143721232_143721232_G_A 1451R>W Substitution 32% coding Nonsynonymous PGDX11T C1orf183 chromosome 1 open reading frame 183 CCDS841.1 chr1_112071336_112071336_C_T 224R>Q Substitution 22% coding PGDX11T CMYA5 cardiomyopathy associated 5 NM_153610 chr5_79063455_79063455_C_A 1037C>X Substitution Nonsense 34% Nonsynonymous PGDX11T CNR1 cannabinoid receptor 1 (brain) CCDS5015.1 chr6_88911646_88911646_C_T 23V>M Substitution 35% coding Nonsynonymous PGDX11T COL4A4 collagen; type IV; alpha 4 CCDS42828.1 chr2_227681807_227681807_G_A 227R>C Substitution 20% coding Nonsynonymous PGDX11T CYBASC3 cytochrome b; ascorbate dependent 3 CCDS8004.1 chr11_60877136_60877136_G_A 149R>C Substitution 34% coding Nonsynonymous PGDX11T DYRK3 dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 3 CCDS30999.1 chr1_204888091_204888091_G_A 309V>I Substitution 44% coding Nonsynonymous PGDX11T ELMO1 engulfment and cell motility 1 CCDS5449.1 chr7_37239295_37239295_G_A 160T>M Substitution 24% coding Nonsynonymous PGDX11T FAM83H family with sequence similarity 83; member H CCDS6410.2 chr8_144884473_144884473_C_A 90G>C Substitution 53% coding Nonsynonymous PGDX11T KPTN kaptin (actin binding protein)