The Development and Improvement of Instructions
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Drug Development for the Irritable Bowel Syndrome: Current Challenges and Future Perspectives
REVIEW ARTICLE published: 01 February 2013 doi: 10.3389/fphar.2013.00007 Drug development for the irritable bowel syndrome: current challenges and future perspectives Fabrizio De Ponti* Department of Medical and Surgical Sciences, University of Bologna, Bologna, Italy Edited by: Medications are frequently used for the treatment of patients with the irritable bowel syn- Angelo A. Izzo, University of Naples drome (IBS), although their actual benefit is often debated. In fact, the recent progress in Federico II, Italy our understanding of the pathophysiology of IBS, accompanied by a large number of preclin- Reviewed by: Elisabetta Barocelli, University of ical and clinical studies of new drugs, has not been matched by a significant improvement Parma, Italy of the armamentarium of medications available to treat IBS. The aim of this review is to Raffaele Capasso, University of outline the current challenges in drug development for IBS, taking advantage of what we Naples Federico II, Italy have learnt through the Rome process (Rome I, Rome II, and Rome III). The key questions *Correspondence: that will be addressed are: (a) do we still believe in the “magic bullet,” i.e., a very selective Fabrizio De Ponti, Pharmacology Unit, Department of Medical and Surgical drug displaying a single receptor mechanism capable of controlling IBS symptoms? (b) IBS Sciences, University of Bologna, Via is a “functional disorder” where complex neuroimmune and brain-gut interactions occur Irnerio, 48, 40126 Bologna, Italy. and minimal inflammation is often documented: -
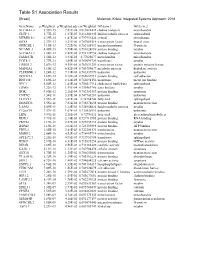
Table S1 Association Results
GeneName p.Weighted p.Weighted.adj cor.Weighted GO.term.1 GO.term.2 SLC44A1.2 3.55E-15 7.81E-08 0.819822422 choline transport mitochondria GLTP.1 5.77E-15 1.27E-07 0.816302445 lipid metabolic process sphingolipid MTMR10.1 6.39E-14 1.41E-06 0.797951424 cytosol phosphatase SOX8 2.37E-13 5.21E-06 0.787085514 transcription factor neural crest GPRC5B.1 4.19E-13 9.22E-06 0.782154937 integral membrane G-protein NCAM1.3 4.45E-13 9.79E-06 0.781624896 protein binding myelin SLC44A1.1 1.28E-12 2.82E-05 0.772132954 choline transport mitochondria FAM107B 1.56E-12 3.43E-05 0.77025677 mitochondria mitochondria UGT8.1 1.77E-12 3.89E-05 0.769099729 transferase myelin ERBB3.2 2.07E-12 4.55E-05 0.767631259 transcription factor protein tyrosine kinase MAN2A1 3.10E-12 6.82E-05 0.763759677 metabolic process hydrolase activity PLEKHH1.1 3.24E-12 7.13E-05 0.763337879 unknown unknown DOCK5.3 3.69E-12 8.12E-05 0.762087913 protein binding cell adhesion RNF130 3.69E-12 8.12E-05 0.762094156 membrane metal ion binding NPC1 6.50E-12 1.43E-04 0.756517114 cholesterol trafficking sphingolipid ERMN 7.22E-12 1.59E-04 0.755469786 actin binding myelin BOK 9.80E-12 2.16E-04 0.752383357 protein binding apoptosis CNTN2 1.54E-11 3.39E-04 0.747743281 unknown unknown ELOVL1 1.55E-11 3.41E-04 0.74764744 fatty acid sphingolipid DBNDD2 3.55E-11 7.81E-04 0.738878658 protein binding neuron projection LASS2 5.09E-11 1.12E-03 0.734954024 lipid metabolic process myelin C12orf34 7.57E-11 1.67E-03 0.730528911 unknown unknown LIPA 9.59E-11 2.11E-03 0.72786111 fatty acid glycerolipid metabolic -

Genetic Variation Across the Human Olfactory Receptor Repertoire Alters Odor Perception
bioRxiv preprint doi: https://doi.org/10.1101/212431; this version posted November 1, 2017. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. Genetic variation across the human olfactory receptor repertoire alters odor perception Casey Trimmer1,*, Andreas Keller2, Nicolle R. Murphy1, Lindsey L. Snyder1, Jason R. Willer3, Maira Nagai4,5, Nicholas Katsanis3, Leslie B. Vosshall2,6,7, Hiroaki Matsunami4,8, and Joel D. Mainland1,9 1Monell Chemical Senses Center, Philadelphia, Pennsylvania, USA 2Laboratory of Neurogenetics and Behavior, The Rockefeller University, New York, New York, USA 3Center for Human Disease Modeling, Duke University Medical Center, Durham, North Carolina, USA 4Department of Molecular Genetics and Microbiology, Duke University Medical Center, Durham, North Carolina, USA 5Department of Biochemistry, University of Sao Paulo, Sao Paulo, Brazil 6Howard Hughes Medical Institute, New York, New York, USA 7Kavli Neural Systems Institute, New York, New York, USA 8Department of Neurobiology and Duke Institute for Brain Sciences, Duke University Medical Center, Durham, North Carolina, USA 9Department of Neuroscience, University of Pennsylvania School of Medicine, Philadelphia, Pennsylvania, USA *[email protected] ABSTRACT The human olfactory receptor repertoire is characterized by an abundance of genetic variation that affects receptor response, but the perceptual effects of this variation are unclear. To address this issue, we sequenced the OR repertoire in 332 individuals and examined the relationship between genetic variation and 276 olfactory phenotypes, including the perceived intensity and pleasantness of 68 odorants at two concentrations, detection thresholds of three odorants, and general olfactory acuity. -

An Overview of the Role of Hdacs in Cancer Immunotherapy
International Journal of Molecular Sciences Review Immunoepigenetics Combination Therapies: An Overview of the Role of HDACs in Cancer Immunotherapy Debarati Banik, Sara Moufarrij and Alejandro Villagra * Department of Biochemistry and Molecular Medicine, School of Medicine and Health Sciences, The George Washington University, 800 22nd St NW, Suite 8880, Washington, DC 20052, USA; [email protected] (D.B.); [email protected] (S.M.) * Correspondence: [email protected]; Tel.: +(202)-994-9547 Received: 22 March 2019; Accepted: 28 April 2019; Published: 7 May 2019 Abstract: Long-standing efforts to identify the multifaceted roles of histone deacetylase inhibitors (HDACis) have positioned these agents as promising drug candidates in combatting cancer, autoimmune, neurodegenerative, and infectious diseases. The same has also encouraged the evaluation of multiple HDACi candidates in preclinical studies in cancer and other diseases as well as the FDA-approval towards clinical use for specific agents. In this review, we have discussed how the efficacy of immunotherapy can be leveraged by combining it with HDACis. We have also included a brief overview of the classification of HDACis as well as their various roles in physiological and pathophysiological scenarios to target key cellular processes promoting the initiation, establishment, and progression of cancer. Given the critical role of the tumor microenvironment (TME) towards the outcome of anticancer therapies, we have also discussed the effect of HDACis on different components of the TME. We then have gradually progressed into examples of specific pan-HDACis, class I HDACi, and selective HDACis that either have been incorporated into clinical trials or show promising preclinical effects for future consideration. -

PDF Datasheet
Product Datasheet C9orf85 Overexpression Lysate NBL1-08604 Unit Size: 0.1 mg Store at -80C. Avoid freeze-thaw cycles. Protocols, Publications, Related Products, Reviews, Research Tools and Images at: www.novusbio.com/NBL1-08604 Updated 3/17/2020 v.20.1 Earn rewards for product reviews and publications. Submit a publication at www.novusbio.com/publications Submit a review at www.novusbio.com/reviews/destination/NBL1-08604 Page 1 of 2 v.20.1 Updated 3/17/2020 NBL1-08604 C9orf85 Overexpression Lysate Product Information Unit Size 0.1 mg Concentration The exact concentration of the protein of interest cannot be determined for overexpression lysates. Please contact technical support for more information. Storage Store at -80C. Avoid freeze-thaw cycles. Buffer RIPA buffer Target Molecular Weight 18.1 kDa Product Description Description Transient overexpression lysate of chromosome 9 open reading frame 85 (C9orf85) The lysate was created in HEK293T cells, using Plasmid ID RC208807 and based on accession number NM_182505. The protein contains a C- MYC/DDK Tag. Gene ID 138241 Gene Symbol C9ORF85 Species Human Notes HEK293T cells in 10-cm dishes were transiently transfected with a non-lipid polymer transfection reagent specially designed and manufactured for large volume DNA transfection. Transfected cells were cultured for 48hrs before collection. The cells were lysed in modified RIPA buffer (25mM Tris-HCl pH7.6, 150mM NaCl, 1% NP-40, 1mM EDTA, 1xProteinase inhibitor cocktail mix, 1mM PMSF and 1mM Na3VO4, and then centrifuged to clarify the lysate. Protein concentration was measured by BCA protein assay kit.This product is manufactured by and sold under license from OriGene Technologies and its use is limited solely for research purposes. -

Table 2. Significant
Table 2. Significant (Q < 0.05 and |d | > 0.5) transcripts from the meta-analysis Gene Chr Mb Gene Name Affy ProbeSet cDNA_IDs d HAP/LAP d HAP/LAP d d IS Average d Ztest P values Q-value Symbol ID (study #5) 1 2 STS B2m 2 122 beta-2 microglobulin 1452428_a_at AI848245 1.75334941 4 3.2 4 3.2316485 1.07398E-09 5.69E-08 Man2b1 8 84.4 mannosidase 2, alpha B1 1416340_a_at H4049B01 3.75722111 3.87309653 2.1 1.6 2.84852656 5.32443E-07 1.58E-05 1110032A03Rik 9 50.9 RIKEN cDNA 1110032A03 gene 1417211_a_at H4035E05 4 1.66015788 4 1.7 2.82772795 2.94266E-05 0.000527 NA 9 48.5 --- 1456111_at 3.43701477 1.85785922 4 2 2.8237185 9.97969E-08 3.48E-06 Scn4b 9 45.3 Sodium channel, type IV, beta 1434008_at AI844796 3.79536664 1.63774235 3.3 2.3 2.75319499 1.48057E-08 6.21E-07 polypeptide Gadd45gip1 8 84.1 RIKEN cDNA 2310040G17 gene 1417619_at 4 3.38875643 1.4 2 2.69163229 8.84279E-06 0.0001904 BC056474 15 12.1 Mus musculus cDNA clone 1424117_at H3030A06 3.95752801 2.42838452 1.9 2.2 2.62132809 1.3344E-08 5.66E-07 MGC:67360 IMAGE:6823629, complete cds NA 4 153 guanine nucleotide binding protein, 1454696_at -3.46081884 -4 -1.3 -1.6 -2.6026947 8.58458E-05 0.0012617 beta 1 Gnb1 4 153 guanine nucleotide binding protein, 1417432_a_at H3094D02 -3.13334396 -4 -1.6 -1.7 -2.5946297 1.04542E-05 0.0002202 beta 1 Gadd45gip1 8 84.1 RAD23a homolog (S. -

United States Patent (10) Patent No.: US 8,969,514 B2 Shailubhai (45) Date of Patent: Mar
USOO896.9514B2 (12) United States Patent (10) Patent No.: US 8,969,514 B2 Shailubhai (45) Date of Patent: Mar. 3, 2015 (54) AGONISTS OF GUANYLATECYCLASE 5,879.656 A 3, 1999 Waldman USEFUL FOR THE TREATMENT OF 36; A 6. 3: Watts tal HYPERCHOLESTEROLEMIA, 6,060,037- W - A 5, 2000 Waldmlegand et al. ATHEROSCLEROSIS, CORONARY HEART 6,235,782 B1 5/2001 NEW et al. DISEASE, GALLSTONE, OBESITY AND 7,041,786 B2 * 5/2006 Shailubhai et al. ........... 530.317 OTHER CARDOVASCULAR DISEASES 2002fOO78683 A1 6/2002 Katayama et al. 2002/O12817.6 A1 9/2002 Forssmann et al. (75) Inventor: Kunwar Shailubhai, Audubon, PA (US) 2003,2002/0143015 OO73628 A1 10/20024, 2003 ShaubhaiFryburg et al. 2005, OO16244 A1 1/2005 H 11 (73) Assignee: Synergy Pharmaceuticals, Inc., New 2005, OO32684 A1 2/2005 Syer York, NY (US) 2005/0267.197 A1 12/2005 Berlin 2006, OO86653 A1 4, 2006 St. Germain (*) Notice: Subject to any disclaimer, the term of this 299;s: A. 299; NS et al. patent is extended or adjusted under 35 2008/0137318 A1 6/2008 Rangarajetal.O U.S.C. 154(b) by 742 days. 2008. O151257 A1 6/2008 Yasuda et al. 2012/O196797 A1 8, 2012 Currie et al. (21) Appl. No.: 12/630,654 FOREIGN PATENT DOCUMENTS (22) Filed: Dec. 3, 2009 DE 19744O27 4f1999 (65) Prior Publication Data WO WO-8805306 T 1988 WO WO99,26567 A1 6, 1999 US 2010/O152118A1 Jun. 17, 2010 WO WO-0 125266 A1 4, 2001 WO WO-02062369 A2 8, 2002 Related U.S. -
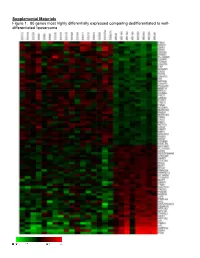
Supplemental Materials Figure 1. 80 Genes Most Highly Differentially Expressed Comparing Dedifferentiated to Well- Differentiated Liposarcoma
Supplemental Materials Figure 1. 80 genes most highly differentially expressed comparing dedifferentiated to well- differentiated liposarcoma Figure 2. Validation of microa CDC2, RACGAP1, FGFR-2, MAD2 and CITED1 200 CDK4 by Liposarcoma Subtype 16 0 12 0 80 Expression40 Level 0 Nl Fat rray data by quantitative RT-P Well Diff 16 0 p16 by Liposarcoma Subtype Dediff 12 0 80 Myxoid Expression40 Level Round Cell 0 12 0 MDM2 by Liposarcoma Subtype 10 0 Pleomorphic Nl Fat 80 60 40 Expression Level 20 RACGAP1 by LiposarcomaWell Diff Subtype 0 70 CR for genes CDK4, MDM2, p16, 60 Dediff 50 Nl Fat 40 30 Myxoid Expression20 Level Well Diff 10 50 0 Round Cell CDC2 by Liposarcoma Subtype Dediff 40 Nl Fat Pleomorphic 30 Myxoid 20 Expression Level Well Diff 10 MAD2L1 by Liposarcoma Subtype Round Cell 60 0 50 Dediff Pleomorphic 40 Nl Fat 30 Myxoid 20 Expression Level Well Diff 10 FGFR-2 by Liposarcoma Subtype 0 Round Cell 200 16 0 Dediff Nl Fat Pleomorphic 12 0 Myxoid 80 Expression Level Well Diff 40 Round Cell 0 Dediff Pleomorphic Nl Fat Myxoid Well Diff CITED1 by Liposarcoma Subtype Round Cell 10 0 80 Dediff Pleomorphic 60 40 Myxoid Expression Level 20 0 Round Cell Nl Fat Pleomorphic Well Diff Dediff Myxoid Round Cell Pleomorphic Table 1. 142 Gene classifier for liposarcoma subtype The genes used for each pair wise subtype comparison are grouped together. The flag column indicates which genes are unique to each subtype comparison. The values show the mean expression levels (actually the mean of the log expression levels was computed and than transformed back to absolute expression level). -

Dual Proteome-Scale Networks Reveal Cell-Specific Remodeling of the Human Interactome
bioRxiv preprint doi: https://doi.org/10.1101/2020.01.19.905109; this version posted January 19, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Dual Proteome-scale Networks Reveal Cell-specific Remodeling of the Human Interactome Edward L. Huttlin1*, Raphael J. Bruckner1,3, Jose Navarrete-Perea1, Joe R. Cannon1,4, Kurt Baltier1,5, Fana Gebreab1, Melanie P. Gygi1, Alexandra Thornock1, Gabriela Zarraga1,6, Stanley Tam1,7, John Szpyt1, Alexandra Panov1, Hannah Parzen1,8, Sipei Fu1, Arvene Golbazi1, Eila Maenpaa1, Keegan Stricker1, Sanjukta Guha Thakurta1, Ramin Rad1, Joshua Pan2, David P. Nusinow1, Joao A. Paulo1, Devin K. Schweppe1, Laura Pontano Vaites1, J. Wade Harper1*, Steven P. Gygi1*# 1Department of Cell Biology, Harvard Medical School, Boston, MA, 02115, USA. 2Broad Institute, Cambridge, MA, 02142, USA. 3Present address: ICCB-Longwood Screening Facility, Harvard Medical School, Boston, MA, 02115, USA. 4Present address: Merck, West Point, PA, 19486, USA. 5Present address: IQ Proteomics, Cambridge, MA, 02139, USA. 6Present address: Vor Biopharma, Cambridge, MA, 02142, USA. 7Present address: Rubius Therapeutics, Cambridge, MA, 02139, USA. 8Present address: RPS North America, South Kingstown, RI, 02879, USA. *Correspondence: [email protected] (E.L.H.), [email protected] (J.W.H.), [email protected] (S.P.G.) #Lead Contact: [email protected] bioRxiv preprint doi: https://doi.org/10.1101/2020.01.19.905109; this version posted January 19, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Transcriptional Control of Tissue-Resident Memory T Cell Generation
Transcriptional control of tissue-resident memory T cell generation Filip Cvetkovski Submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy in the Graduate School of Arts and Sciences COLUMBIA UNIVERSITY 2019 © 2019 Filip Cvetkovski All rights reserved ABSTRACT Transcriptional control of tissue-resident memory T cell generation Filip Cvetkovski Tissue-resident memory T cells (TRM) are a non-circulating subset of memory that are maintained at sites of pathogen entry and mediate optimal protection against reinfection. Lung TRM can be generated in response to respiratory infection or vaccination, however, the molecular pathways involved in CD4+TRM establishment have not been defined. Here, we performed transcriptional profiling of influenza-specific lung CD4+TRM following influenza infection to identify pathways implicated in CD4+TRM generation and homeostasis. Lung CD4+TRM displayed a unique transcriptional profile distinct from spleen memory, including up-regulation of a gene network induced by the transcription factor IRF4, a known regulator of effector T cell differentiation. In addition, the gene expression profile of lung CD4+TRM was enriched in gene sets previously described in tissue-resident regulatory T cells. Up-regulation of immunomodulatory molecules such as CTLA-4, PD-1, and ICOS, suggested a potential regulatory role for CD4+TRM in tissues. Using loss-of-function genetic experiments in mice, we demonstrate that IRF4 is required for the generation of lung-localized pathogen-specific effector CD4+T cells during acute influenza infection. Influenza-specific IRF4−/− T cells failed to fully express CD44, and maintained high levels of CD62L compared to wild type, suggesting a defect in complete differentiation into lung-tropic effector T cells. -

Technical Note, Appendix: an Analysis of Blood Processing Methods to Prepare Samples for Genechip® Expression Profiling (Pdf, 1
Appendix 1: Signature genes for different blood cell types. Blood Cell Type Source Probe Set Description Symbol Blood Cell Type Source Probe Set Description Symbol Fraction ID Fraction ID Mono- Lympho- GSK 203547_at CD4 antigen (p55) CD4 Whitney et al. 209813_x_at T cell receptor TRG nuclear cytes gamma locus cells Whitney et al. 209995_s_at T-cell leukemia/ TCL1A Whitney et al. 203104_at colony stimulating CSF1R lymphoma 1A factor 1 receptor, Whitney et al. 210164_at granzyme B GZMB formerly McDonough (granzyme 2, feline sarcoma viral cytotoxic T-lymphocyte- (v-fms) oncogene associated serine homolog esterase 1) Whitney et al. 203290_at major histocompatibility HLA-DQA1 Whitney et al. 210321_at similar to granzyme B CTLA1 complex, class II, (granzyme 2, cytotoxic DQ alpha 1 T-lymphocyte-associated Whitney et al. 203413_at NEL-like 2 (chicken) NELL2 serine esterase 1) Whitney et al. 203828_s_at natural killer cell NK4 (H. sapiens) transcript 4 Whitney et al. 212827_at immunoglobulin heavy IGHM Whitney et al. 203932_at major histocompatibility HLA-DMB constant mu complex, class II, Whitney et al. 212998_x_at major histocompatibility HLA-DQB1 DM beta complex, class II, Whitney et al. 204655_at chemokine (C-C motif) CCL5 DQ beta 1 ligand 5 Whitney et al. 212999_x_at major histocompatibility HLA-DQB Whitney et al. 204661_at CDW52 antigen CDW52 complex, class II, (CAMPATH-1 antigen) DQ beta 1 Whitney et al. 205049_s_at CD79A antigen CD79A Whitney et al. 213193_x_at T cell receptor beta locus TRB (immunoglobulin- Whitney et al. 213425_at Homo sapiens cDNA associated alpha) FLJ11441 fis, clone Whitney et al. 205291_at interleukin 2 receptor, IL2RB HEMBA1001323, beta mRNA sequence Whitney et al.