Molecular and Morphological Identification of an Uncommon Centrolophid Fish
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Biology and Conservation of Xantus's Murrelet
SYMPOSIUM BIOLOGY AND CONSERVATION OF XANTUS’S MURRELET Guest editors Harry R. Carter, Spencer G. Sealy, Esther E. Burkett and John F. Piatt Xantus’s Murrelet chicks shortly before nest departure at Lava Bench, Cave #1, Anacapa Island, California, 28 May 2004 (Photo by D.L. Whitworth). Xantus'sCarter et Murrelet al.: Biology Synthliboramphus and conservation H ypoleucusof Xantus’s Symposium Murrelet 81 BIOLOGY AND CONSERVATION OF XANTUS’S MURRELET: DISCOVERY, TAXONOMY AND DISTRIBUTION HARRY R. CARTER1, SPENCER G. SEALY2, ESTHER E. BURKETT3 & JOHN F. PIATT4 1Carter Biological Consulting, 1015 Hampshire Road, Victoria, British Columbia, V8S 4S8, Canada ([email protected]) 2Department of Zoology, University of Manitoba, Winnipeg, Manitoba, R3T 2N2, Canada 3California Department of Fish and Game, Habitat Conservation and Planning Branch, 1416 Ninth Street, Sacramento, California, 95814, USA 4US Geological Survey, Alaska Science Center, 1011 East Tudor Road, Anchorage, Alaska, 99503, USA Received 28 October 2005, accepted 12 January 2006 SUMMARY CARTER, H.R., SEALY, S.G., BURKETT, E.E. & PIATT, J.F. 2005. Biology and conservation of Xantus’s Murrelet: discovery, taxonomy, and distribution. Marine Ornithology 33: 81–87. The biology of Xantus’s Murrelets Synthliboramphus hypoleucus is similar in many respects to better-studied Ancient Murrelets S. antiquus, especially regarding morphology and the species’ precocial mode of post-hatching development. It nests mainly in rock crevices but also under shrubs on islands in southern California, United States, and northwestern Baja California, Mexico (27°N to 34°N). The species was discovered in 1859 by János Xántus. Two subspecies (S. h. hypoleucus and S. h. scrippsi) are recognized that show limited evidence of interbreeding. -

A Practical Handbook for Determining the Ages of Gulf of Mexico And
A Practical Handbook for Determining the Ages of Gulf of Mexico and Atlantic Coast Fishes THIRD EDITION GSMFC No. 300 NOVEMBER 2020 i Gulf States Marine Fisheries Commission Commissioners and Proxies ALABAMA Senator R.L. “Bret” Allain, II Chris Blankenship, Commissioner State Senator District 21 Alabama Department of Conservation Franklin, Louisiana and Natural Resources John Roussel Montgomery, Alabama Zachary, Louisiana Representative Chris Pringle Mobile, Alabama MISSISSIPPI Chris Nelson Joe Spraggins, Executive Director Bon Secour Fisheries, Inc. Mississippi Department of Marine Bon Secour, Alabama Resources Biloxi, Mississippi FLORIDA Read Hendon Eric Sutton, Executive Director USM/Gulf Coast Research Laboratory Florida Fish and Wildlife Ocean Springs, Mississippi Conservation Commission Tallahassee, Florida TEXAS Representative Jay Trumbull Carter Smith, Executive Director Tallahassee, Florida Texas Parks and Wildlife Department Austin, Texas LOUISIANA Doug Boyd Jack Montoucet, Secretary Boerne, Texas Louisiana Department of Wildlife and Fisheries Baton Rouge, Louisiana GSMFC Staff ASMFC Staff Mr. David M. Donaldson Mr. Bob Beal Executive Director Executive Director Mr. Steven J. VanderKooy Mr. Jeffrey Kipp IJF Program Coordinator Stock Assessment Scientist Ms. Debora McIntyre Dr. Kristen Anstead IJF Staff Assistant Fisheries Scientist ii A Practical Handbook for Determining the Ages of Gulf of Mexico and Atlantic Coast Fishes Third Edition Edited by Steve VanderKooy Jessica Carroll Scott Elzey Jessica Gilmore Jeffrey Kipp Gulf States Marine Fisheries Commission 2404 Government St Ocean Springs, MS 39564 and Atlantic States Marine Fisheries Commission 1050 N. Highland Street Suite 200 A-N Arlington, VA 22201 Publication Number 300 November 2020 A publication of the Gulf States Marine Fisheries Commission pursuant to National Oceanic and Atmospheric Administration Award Number NA15NMF4070076 and NA15NMF4720399. -

Pacific Plate Biogeography, with Special Reference to Shorefishes
Pacific Plate Biogeography, with Special Reference to Shorefishes VICTOR G. SPRINGER m SMITHSONIAN CONTRIBUTIONS TO ZOOLOGY • NUMBER 367 SERIES PUBLICATIONS OF THE SMITHSONIAN INSTITUTION Emphasis upon publication as a means of "diffusing knowledge" was expressed by the first Secretary of the Smithsonian. In his formal plan for the Institution, Joseph Henry outlined a program that included the following statement: "It is proposed to publish a series of reports, giving an account of the new discoveries in science, and of the changes made from year to year in all branches of knowledge." This theme of basic research has been adhered to through the years by thousands of titles issued in series publications under the Smithsonian imprint, commencing with Smithsonian Contributions to Knowledge in 1848 and continuing with the following active series: Smithsonian Contributions to Anthropology Smithsonian Contributions to Astrophysics Smithsonian Contributions to Botany Smithsonian Contributions to the Earth Sciences Smithsonian Contributions to the Marine Sciences Smithsonian Contributions to Paleobiology Smithsonian Contributions to Zoo/ogy Smithsonian Studies in Air and Space Smithsonian Studies in History and Technology In these series, the Institution publishes small papers and full-scale monographs that report the research and collections of its various museums and bureaux or of professional colleagues in the world cf science and scholarship. The publications are distributed by mailing lists to libraries, universities, and similar institutions throughout the world. Papers or monographs submitted for series publication are received by the Smithsonian Institution Press, subject to its own review for format and style, only through departments of the various Smithsonian museums or bureaux, where the manuscripts are given substantive review. -

CHECKLIST and BIOGEOGRAPHY of FISHES from GUADALUPE ISLAND, WESTERN MEXICO Héctor Reyes-Bonilla, Arturo Ayala-Bocos, Luis E
ReyeS-BONIllA eT Al: CheCklIST AND BIOgeOgRAphy Of fISheS fROm gUADAlUpe ISlAND CalCOfI Rep., Vol. 51, 2010 CHECKLIST AND BIOGEOGRAPHY OF FISHES FROM GUADALUPE ISLAND, WESTERN MEXICO Héctor REyES-BONILLA, Arturo AyALA-BOCOS, LUIS E. Calderon-AGUILERA SAúL GONzáLEz-Romero, ISRAEL SáNCHEz-ALCántara Centro de Investigación Científica y de Educación Superior de Ensenada AND MARIANA Walther MENDOzA Carretera Tijuana - Ensenada # 3918, zona Playitas, C.P. 22860 Universidad Autónoma de Baja California Sur Ensenada, B.C., México Departamento de Biología Marina Tel: +52 646 1750500, ext. 25257; Fax: +52 646 Apartado postal 19-B, CP 23080 [email protected] La Paz, B.C.S., México. Tel: (612) 123-8800, ext. 4160; Fax: (612) 123-8819 NADIA C. Olivares-BAñUELOS [email protected] Reserva de la Biosfera Isla Guadalupe Comisión Nacional de áreas Naturales Protegidas yULIANA R. BEDOLLA-GUzMáN AND Avenida del Puerto 375, local 30 Arturo RAMíREz-VALDEz Fraccionamiento Playas de Ensenada, C.P. 22880 Universidad Autónoma de Baja California Ensenada, B.C., México Facultad de Ciencias Marinas, Instituto de Investigaciones Oceanológicas Universidad Autónoma de Baja California, Carr. Tijuana-Ensenada km. 107, Apartado postal 453, C.P. 22890 Ensenada, B.C., México ABSTRACT recognized the biological and ecological significance of Guadalupe Island, off Baja California, México, is Guadalupe Island, and declared it a Biosphere Reserve an important fishing area which also harbors high (SEMARNAT 2005). marine biodiversity. Based on field data, literature Guadalupe Island is isolated, far away from the main- reviews, and scientific collection records, we pres- land and has limited logistic facilities to conduct scien- ent a comprehensive checklist of the local fish fauna, tific studies. -

Table S1.Xlsx
Bone type Bone type Taxonomy Order/series Family Valid binomial Outdated binomial Notes Reference(s) (skeletal bone) (scales) Actinopterygii Incertae sedis Incertae sedis Incertae sedis †Birgeria stensioei cellular this study †Birgeria groenlandica cellular Ørvig, 1978 †Eurynotus crenatus cellular Goodrich, 1907; Schultze, 2016 †Mimipiscis toombsi †Mimia toombsi cellular Richter & Smith, 1995 †Moythomasia sp. cellular cellular Sire et al., 2009; Schultze, 2016 †Cheirolepidiformes †Cheirolepididae †Cheirolepis canadensis cellular cellular Goodrich, 1907; Sire et al., 2009; Zylberberg et al., 2016; Meunier et al. 2018a; this study Cladistia Polypteriformes Polypteridae †Bawitius sp. cellular Meunier et al., 2016 †Dajetella sudamericana cellular cellular Gayet & Meunier, 1992 Erpetoichthys calabaricus Calamoichthys sp. cellular Moss, 1961a; this study †Pollia suarezi cellular cellular Meunier & Gayet, 1996 Polypterus bichir cellular cellular Kölliker, 1859; Stéphan, 1900; Goodrich, 1907; Ørvig, 1978 Polypterus delhezi cellular this study Polypterus ornatipinnis cellular Totland et al., 2011 Polypterus senegalus cellular Sire et al., 2009 Polypterus sp. cellular Moss, 1961a †Scanilepis sp. cellular Sire et al., 2009 †Scanilepis dubia cellular cellular Ørvig, 1978 †Saurichthyiformes †Saurichthyidae †Saurichthys sp. cellular Scheyer et al., 2014 Chondrostei †Chondrosteiformes †Chondrosteidae †Chondrosteus acipenseroides cellular this study Acipenseriformes Acipenseridae Acipenser baerii cellular Leprévost et al., 2017 Acipenser gueldenstaedtii -

Vulnerable Marine Ecosystems – Processes and Practices in the High Seas Vulnerable Marine Ecosystems Processes and Practices in the High Seas
ISSN 2070-7010 FAO 595 FISHERIES AND AQUACULTURE TECHNICAL PAPER 595 Vulnerable marine ecosystems – Processes and practices in the high seas Vulnerable marine ecosystems Processes and practices in the high seas This publication, Vulnerable Marine Ecosystems: processes and practices in the high seas, provides regional fisheries management bodies, States, and other interested parties with a summary of existing regional measures to protect vulnerable marine ecosystems from significant adverse impacts caused by deep-sea fisheries using bottom contact gears in the high seas. This publication compiles and summarizes information on the processes and practices of the regional fishery management bodies, with mandates to manage deep-sea fisheries in the high seas, to protect vulnerable marine ecosystems. ISBN 978-92-5-109340-5 ISSN 2070-7010 FAO 9 789251 093405 I5952E/2/03.17 Cover photo credits: Photo descriptions clockwise from top-left: Acanthagorgia spp., Paragorgia arborea, Vase sponges (images courtesy of Fisheries and Oceans, Canada); and Callogorgia spp. (image courtesy of Kirsty Kemp, the Zoological Society of London). FAO FISHERIES AND Vulnerable marine ecosystems AQUACULTURE TECHNICAL Processes and practices in the high seas PAPER 595 Edited by Anthony Thompson FAO Consultant Rome, Italy Jessica Sanders Fisheries Officer FAO Fisheries and Aquaculture Department Rome, Italy Merete Tandstad Fisheries Resources Officer FAO Fisheries and Aquaculture Department Rome, Italy Fabio Carocci Fishery Information Assistant FAO Fisheries and Aquaculture Department Rome, Italy and Jessica Fuller FAO Consultant Rome, Italy FOOD AND AGRICULTURE ORGANIZATION OF THE UNITED NATIONS Rome, 2016 The designations employed and the presentation of material in this information product do not imply the expression of any opinion whatsoever on the part of the Food and Agriculture Organization of the United Nations (FAO) concerning the legal or development status of any country, territory, city or area or of its authorities, or concerning the delimitation of its frontiers or boundaries. -

© Iccat, 2007
A5 By-catch Species APPENDIX 5: BY-CATCH SPECIES A.5 By-catch species By-catch is the unintentional/incidental capture of non-target species during fishing operations. Different types of fisheries have different types and levels of by-catch, depending on the gear used, the time, area and depth fished, etc. Article IV of the Convention states: "the Commission shall be responsible for the study of the population of tuna and tuna-like fishes (the Scombriformes with the exception of Trichiuridae and Gempylidae and the genus Scomber) and such other species of fishes exploited in tuna fishing in the Convention area as are not under investigation by another international fishery organization". The following is a list of by-catch species recorded as being ever caught by any major tuna fishery in the Atlantic/Mediterranean. Note that the lists are qualitative and are not indicative of quantity or mortality. Thus, the presence of a species in the lists does not imply that it is caught in significant quantities, or that individuals that are caught necessarily die. Skates and rays Scientific names Common name Code LL GILL PS BB HARP TRAP OTHER Dasyatis centroura Roughtail stingray RDC X Dasyatis violacea Pelagic stingray PLS X X X X Manta birostris Manta ray RMB X X X Mobula hypostoma RMH X Mobula lucasana X Mobula mobular Devil ray RMM X X X X X Myliobatis aquila Common eagle ray MYL X X Pteuromylaeus bovinus Bull ray MPO X X Raja fullonica Shagreen ray RJF X Raja straeleni Spotted skate RFL X Rhinoptera spp Cownose ray X Torpedo nobiliana Torpedo -

Updated Checklist of Marine Fishes (Chordata: Craniata) from Portugal and the Proposed Extension of the Portuguese Continental Shelf
European Journal of Taxonomy 73: 1-73 ISSN 2118-9773 http://dx.doi.org/10.5852/ejt.2014.73 www.europeanjournaloftaxonomy.eu 2014 · Carneiro M. et al. This work is licensed under a Creative Commons Attribution 3.0 License. Monograph urn:lsid:zoobank.org:pub:9A5F217D-8E7B-448A-9CAB-2CCC9CC6F857 Updated checklist of marine fishes (Chordata: Craniata) from Portugal and the proposed extension of the Portuguese continental shelf Miguel CARNEIRO1,5, Rogélia MARTINS2,6, Monica LANDI*,3,7 & Filipe O. COSTA4,8 1,2 DIV-RP (Modelling and Management Fishery Resources Division), Instituto Português do Mar e da Atmosfera, Av. Brasilia 1449-006 Lisboa, Portugal. E-mail: [email protected], [email protected] 3,4 CBMA (Centre of Molecular and Environmental Biology), Department of Biology, University of Minho, Campus de Gualtar, 4710-057 Braga, Portugal. E-mail: [email protected], [email protected] * corresponding author: [email protected] 5 urn:lsid:zoobank.org:author:90A98A50-327E-4648-9DCE-75709C7A2472 6 urn:lsid:zoobank.org:author:1EB6DE00-9E91-407C-B7C4-34F31F29FD88 7 urn:lsid:zoobank.org:author:6D3AC760-77F2-4CFA-B5C7-665CB07F4CEB 8 urn:lsid:zoobank.org:author:48E53CF3-71C8-403C-BECD-10B20B3C15B4 Abstract. The study of the Portuguese marine ichthyofauna has a long historical tradition, rooted back in the 18th Century. Here we present an annotated checklist of the marine fishes from Portuguese waters, including the area encompassed by the proposed extension of the Portuguese continental shelf and the Economic Exclusive Zone (EEZ). The list is based on historical literature records and taxon occurrence data obtained from natural history collections, together with new revisions and occurrences. -

A Dissertation Entitled Evolution, Systematics
A Dissertation Entitled Evolution, systematics, and phylogeography of Ponto-Caspian gobies (Benthophilinae: Gobiidae: Teleostei) By Matthew E. Neilson Submitted as partial fulfillment of the requirements for The Doctor of Philosophy Degree in Biology (Ecology) ____________________________________ Adviser: Dr. Carol A. Stepien ____________________________________ Committee Member: Dr. Christine M. Mayer ____________________________________ Committee Member: Dr. Elliot J. Tramer ____________________________________ Committee Member: Dr. David J. Jude ____________________________________ Committee Member: Dr. Juan L. Bouzat ____________________________________ College of Graduate Studies The University of Toledo December 2009 Copyright © 2009 This document is copyrighted material. Under copyright law, no parts of this document may be reproduced without the expressed permission of the author. _______________________________________________________________________ An Abstract of Evolution, systematics, and phylogeography of Ponto-Caspian gobies (Benthophilinae: Gobiidae: Teleostei) Matthew E. Neilson Submitted as partial fulfillment of the requirements for The Doctor of Philosophy Degree in Biology (Ecology) The University of Toledo December 2009 The study of biodiversity, at multiple hierarchical levels, provides insight into the evolutionary history of taxa and provides a framework for understanding patterns in ecology. This is especially poignant in invasion biology, where the prevalence of invasiveness in certain taxonomic groups could -
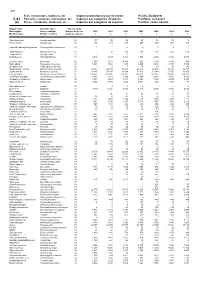
Fish, Crustaceans, Molluscs, Etc Capture Production by Species
478 Fish, crustaceans, molluscs, etc Capture production by species items Pacific, Southwest C-81 Poissons, crustacés, mollusques, etc Captures par catégories d'espèces Pacifique, sud-ouest (a) Peces, crustáceos, moluscos, etc Capturas por categorías de especies Pacífico, sudoccidental English name Scientific name Species group Nom anglais Nom scientifique Groupe d'espèces 2000 2001 2002 2003 2004 2005 2006 Nombre inglés Nombre científico Grupo de especies t t t t t t t Short-finned eel Anguilla australis 22 37 38 28 27 13 10 5 River eels nei Anguilla spp 22 380 313 337 267 209 277 210 Chinook(=Spring=King)salmon Oncorhynchus tshawytscha 23 1 1 0 4 1 2 1 Sand flounders Rhombosolea spp 31 ... 37 204 193 187 437 514 Tonguefishes Cynoglossidae 31 - - 3 - - - - Flatfishes nei Pleuronectiformes 31 2 954 3 234 2 818 3 308 2 980 3 766 3 050 Common mora Mora moro 32 1 358 1 211 1 308 1 234 1 403 1 154 986 Red codling Pseudophycis bachus 32 5 364 4 526 4 443 8 265 9 540 8 165 5 854 Grenadier cod Tripterophycis gilchristi 32 - 2 7 10 13 13 43 Southern blue whiting Micromesistius australis 32 43 419 49 982 72 203 43 812 26 576 30 304 32 735 Southern hake Merluccius australis 32 16 041 15 188 13 834 22 623 19 344 12 560 12 858 Blue grenadier Macruronus novaezelandiae 32 274 625 247 841 215 302 209 414 147 032 134 145 119 329 Thorntooth grenadier Lepidorhynchus denticulatus 32 3 833 4 783 5 349 5 304 6 341 3 855 4 056 Grenadiers, rattails nei Macrouridae 32 2 394 3 094 3 877 4 253 3 732 2 660 2 848 Gadiformes nei Gadiformes 32 2 853 5 479 3 252 3 281 298 1 217 47 Sea catfishes nei Ariidae 33 .. -

Checklist of Marine Demersal Fishes Captured by the Pair Trawl Fisheries in Southern (RJ-SC) Brazil
Biota Neotropica 19(1): e20170432, 2019 www.scielo.br/bn ISSN 1676-0611 (online edition) Inventory Checklist of marine demersal fishes captured by the pair trawl fisheries in Southern (RJ-SC) Brazil Matheus Marcos Rotundo1,2,3,4 , Evandro Severino-Rodrigues2, Walter Barrella4,5, Miguel Petrere Jun- ior3 & Milena Ramires4,5 1Universidade Santa Cecilia, Acervo Zoológico, R. Oswaldo Cruz, 266, CEP11045-907, Santos, SP, Brasil 2Instituto de Pesca, Programa de Pós-graduação em Aquicultura e Pesca, Santos, SP, Brasil 3Universidade Federal de São Carlos, Programa de Pós-Graduação em Planejamento e Uso de Recursos Renováveis, Rodovia João Leme dos Santos, Km 110, CEP 18052-780, Sorocaba, SP, Brasil 4Universidade Santa Cecília, Programa de Pós-Graduação de Auditoria Ambiental, R. Oswaldo Cruz, 266, CEP11045-907, Santos, SP, Brasil 5Universidade Santa Cecília, Programa de Pós-Graduação em Sustentabilidade de Ecossistemas Costeiros e Marinhos, R. Oswaldo Cruz, 266, CEP11045-907, Santos, SP, Brasil *Corresponding author: Matheus Marcos Rotundo: [email protected] ROTUNDO, M.M., SEVERINO-RODRIGUES, E., BARRELLA, W., PETRERE JUNIOR, M., RAMIRES, M. Checklist of marine demersal fishes captured by the pair trawl fisheries in Southern (RJ-SC) Brazil. Biota Neotropica. 19(1): e20170432. http://dx.doi.org/10.1590/1676-0611-BN-2017-0432 Abstract: Demersal fishery resources are abundant on continental shelves, on the tropical and subtropical coasts, making up a significant part of the marine environment. Marine demersal fishery resources are captured by various fishing methods, often unsustainably, which has led to the depletion of their stocks. In order to inventory the marine demersal ichthyofauna on the Southern Brazilian coast, as well as their conservation status and distribution, this study analyzed the composition and frequency of occurrence of fish captured by pair trawling in 117 fishery fleet landings based in the State of São Paulo between 2005 and 2012. -

Appendices Appendices
APPENDICES APPENDICES APPENDIX 1 – PUBLICATIONS SCIENTIFIC PAPERS Aidoo EN, Ute Mueller U, Hyndes GA, and Ryan Braccini M. 2015. Is a global quantitative KL. 2016. The effects of measurement uncertainty assessment of shark populations warranted? on spatial characterisation of recreational fishing Fisheries, 40: 492–501. catch rates. Fisheries Research 181: 1–13. Braccini M. 2016. Experts have different Andrews KR, Williams AJ, Fernandez-Silva I, perceptions of the management and conservation Newman SJ, Copus JM, Wakefield CB, Randall JE, status of sharks. Annals of Marine Biology and and Bowen BW. 2016. Phylogeny of deepwater Research 3: 1012. snappers (Genus Etelis) reveals a cryptic species pair in the Indo-Pacific and Pleistocene invasion of Braccini M, Aires-da-Silva A, and Taylor I. 2016. the Atlantic. Molecular Phylogenetics and Incorporating movement in the modelling of shark Evolution 100: 361-371. and ray population dynamics: approaches and management implications. Reviews in Fish Biology Bellchambers LM, Gaughan D, Wise B, Jackson G, and Fisheries 26: 13–24. and Fletcher WJ. 2016. Adopting Marine Stewardship Council certification of Western Caputi N, de Lestang S, Reid C, Hesp A, and How J. Australian fisheries at a jurisdictional level: the 2015. Maximum economic yield of the western benefits and challenges. Fisheries Research 183: rock lobster fishery of Western Australia after 609-616. moving from effort to quota control. Marine Policy, 51: 452-464. Bellchambers LM, Fisher EA, Harry AV, and Travaille KL. 2016. Identifying potential risks for Charles A, Westlund L, Bartley DM, Fletcher WJ, Marine Stewardship Council assessment and Garcia S, Govan H, and Sanders J.