2018-Bellii-Phylogeography-Mpe.Pdf
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Rediscovery at the Type Locality and Detection of a New Population 1José L
Offcial journal website: Amphibian & Reptile Conservation amphibian-reptile-conservation.org 13(2) [General Section]: 126–132 (e193). Thorius narismagnus (Amphibia: Plethodontidae): rediscovery at the type locality and detection of a new population 1José L. Aguilar-López, 2,*Paulina García-Bañuelos, 3Eduardo Pineda, and 4Sean M. Rovito 1,2,3Red de Biología y Conservación de Vertebrados, Instituto de Ecología, A.C., Carretera antigua a Coatepec 351, El Haya, Xalapa, Veracruz, MEXICO 4Unidad de Genómica Avanzada (Langebio), Centro de Investigación y de Estudios Avanzados del Instituto Politécnico Nacional, km 9.6 Libramiento Norte Carretera Irapuato-León, Irapuato, Guanajuato CP 36824, MEXICO Abstract.—Of the 42 Critically Endangered species of plethodontid salamanders that occur in Mexico, thirteen have not been reported in more than ten years. Given the lack of reports since 1976, the minute plethodontid salamander Thorius narismagnus is widely considered as missing. However, this report describes the rediscovery of this minute salamander at the type locality (Volcán San Martín), as well as a new locality on Volcán Santa Marta, 28 km southeast of its previously known distribution, both in the Los Tuxtlas region of Veracruz, Mexico. The localities where T. narismagnus has been found are mature forests in a community reserve on Volcán San Martín and a private reserve on Volcán Santa Marta. The presence of maxillary teeth, generally absent in Thorius, are reported here in some T. narismagnus females. Two efforts which may contribute to the conservation of Thorius narismagnus are the preservation of the cloud forests where this species persists, as well as the determination of the presence and possible effect of chytrid fungus in these populations. -

Assessing Environmental Variables Across Plethodontid Salamanders
Lauren Mellenthin1, Erica Baken1, Dr. Dean Adams1 1Iowa State University, Ames, Iowa Stereochilus marginatus Pseudotriton ruber Pseudotriton montanus Gyrinophilus subterraneus Gyrinophilus porphyriticus Gyrinophilus palleucus Gyrinophilus gulolineatus Urspelerpes brucei Eurycea tynerensis Eurycea spelaea Eurycea multiplicata Introduction Eurycea waterlooensis Eurycea rathbuni Materials & Methods Eurycea sosorum Eurycea tridentifera Eurycea pterophila Eurycea neotenes Eurycea nana Eurycea troglodytes Eurycea latitans Eurycea naufragia • Arboreality has evolved at least 5 times within Plethodontid salamanders. [1] Eurycea tonkawae Eurycea chisholmensis Climate Variables & Eurycea quadridigitata Polygons & Point Data Eurycea wallacei Eurycea lucifuga Eurycea longicauda Eurycea guttolineata MAXENT Modeling Eurycea bislineata Eurycea wilderae Eurycea cirrigera • Yet no morphological differences separate arboreal and terrestrial species. [1] Eurycea junaluska Hemidactylium scutatum Batrachoseps robustus Batrachoseps wrighti Batrachoseps campi Batrachoseps attenuatus Batrachoseps pacificus Batrachoseps major Batrachoseps luciae Batrachoseps minor Batrachoseps incognitus • There is minimal range overlap between the two microhabitat types. Batrachoseps gavilanensis Batrachoseps gabrieli Batrachoseps stebbinsi Batrachoseps relictus Batrachoseps simatus Batrachoseps nigriventris Batrachoseps gregarius Preliminary results discovered that 71% of the arboreal species distribution Batrachoseps diabolicus Batrachoseps regius Batrachoseps kawia Dendrotriton -
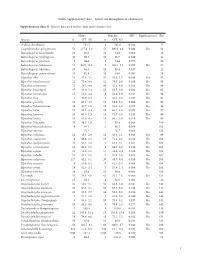
I Online Supplementary Data – Sexual Size Dimorphism in Salamanders
Online Supplementary data – Sexual size dimorphism in salamanders Supplementary data S1. Species data used in this study and references list. Males Females SSD Significant test Ref Species n SVL±SD n SVL±SD Andrias davidianus 2 532.5 8 383.0 -0.280 12 Cryptobranchus alleganiensis 53 277.4±5.2 52 300.9±3.4 0.084 Yes 61 Batrachuperus karlschmidti 10 80.0 10 84.8 0.060 26 Batrachuperus londongensis 20 98.6 10 96.7 -0.019 12 Batrachuperus pinchonii 5 69.6 5 74.6 0.070 26 Batrachuperus taibaiensis 11 92.9±12.1 9 102.1±7.1 0.099 Yes 27 Batrachuperus tibetanus 10 94.5 10 92.8 -0.017 12 Batrachuperus yenyuadensis 10 82.8 10 74.8 -0.096 26 Hynobius abei 24 57.8±2.1 34 55.0±1.2 -0.048 Yes 92 Hynobius amakusaensis 22 75.4±4.8 12 76.5±3.6 0.014 No 93 Hynobius arisanensis 72 54.3±4.8 40 55.2±4.8 0.016 No 94 Hynobius boulengeri 37 83.0±5.4 15 91.5±3.8 0.102 Yes 95 Hynobius formosanus 15 53.0±4.4 8 52.4±3.9 -0.011 No 94 Hynobius fuca 4 50.9±2.8 3 52.8±2.0 0.037 No 94 Hynobius glacialis 12 63.1±4.7 11 58.9±5.2 -0.066 No 94 Hynobius hidamontanus 39 47.7±1.0 15 51.3±1.2 0.075 Yes 96 Hynobius katoi 12 58.4±3.3 10 62.7±1.6 0.073 Yes 97 Hynobius kimurae 20 63.0±1.5 15 72.7±2.0 0.153 Yes 98 Hynobius leechii 70 61.6±4.5 18 66.5±5.9 0.079 Yes 99 Hynobius lichenatus 37 58.5±1.9 2 53.8 -0.080 100 Hynobius maoershanensis 4 86.1 2 80.1 -0.069 101 Hynobius naevius 72.1 76.7 0.063 102 Hynobius nebulosus 14 48.3±2.9 12 50.4±2.1 0.043 Yes 96 Hynobius osumiensis 9 68.4±3.1 15 70.2±3.0 0.026 No 103 Hynobius quelpaertensis 41 52.5±3.8 4 61.3±4.1 0.167 Yes 104 Hynobius -

July to December 2019 (Pdf)
2019 Journal Publications July Adelizzi, R. Portmann, J. van Meter, R. (2019). Effect of Individual and Combined Treatments of Pesticide, Fertilizer, and Salt on Growth and Corticosterone Levels of Larval Southern Leopard Frogs (Lithobates sphenocephala). Archives of Environmental Contamination and Toxicology, 77(1), pp.29-39. https://www.ncbi.nlm.nih.gov/pubmed/31020372 Albecker, M. A. McCoy, M. W. (2019). Local adaptation for enhanced salt tolerance reduces non‐ adaptive plasticity caused by osmotic stress. Evolution, Early View. https://onlinelibrary.wiley.com/doi/abs/10.1111/evo.13798 Alvarez, M. D. V. Fernandez, C. Cove, M. V. (2019). Assessing the role of habitat and species interactions in the population decline and detection bias of Neotropical leaf litter frogs in and around La Selva Biological Station, Costa Rica. Neotropical Biology and Conservation 14(2), pp.143– 156, e37526. https://neotropical.pensoft.net/article/37526/list/11/ Amat, F. Rivera, X. Romano, A. Sotgiu, G. (2019). Sexual dimorphism in the endemic Sardinian cave salamander (Atylodes genei). Folia Zoologica, 68(2), p.61-65. https://bioone.org/journals/Folia-Zoologica/volume-68/issue-2/fozo.047.2019/Sexual-dimorphism- in-the-endemic-Sardinian-cave-salamander-Atylodes-genei/10.25225/fozo.047.2019.short Amézquita, A, Suárez, G. Palacios-Rodríguez, P. Beltrán, I. Rodríguez, C. Barrientos, L. S. Daza, J. M. Mazariegos, L. (2019). A new species of Pristimantis (Anura: Craugastoridae) from the cloud forests of Colombian western Andes. Zootaxa, 4648(3). https://www.biotaxa.org/Zootaxa/article/view/zootaxa.4648.3.8 Arrivillaga, C. Oakley, J. Ebiner, S. (2019). Predation of Scinax ruber (Anura: Hylidae) tadpoles by a fishing spider of the genus Thaumisia (Araneae: Pisauridae) in south-east Peru. -
![Pseudoeurycea Robertsi[I]](https://docslib.b-cdn.net/cover/6530/pseudoeurycea-robertsi-i-2916530.webp)
Pseudoeurycea Robertsi[I]
Natural history of the critically endangered salamander Pseudoeurycea robertsi. Armando Sunny Corresp., Equal first author, 1 , Carmen Caballero-Viñas 2 , Luis Duarte-deJesus 1 , Fabiola Ramírez-Corona 3 , Javier Manjarrez 4 , Giovanny González-Desales 1 , Xareni P. Pacheco 1 , Octavio Monroy-Vilchis 1 , Andrea González- Fernández Corresp. Equal first author, 4 1 Centro de Investigación en Ciencias Biológicas Aplicadas, Universidad Autónoma del Estado de México, Toluca, Estado de México, México 2 Laboratorio de Sistemas Biosustentables, Universidad Autónoma del Estado de México, Toluca, Estado de México, México 3 Departamento de Biología Evolutiva, Facultad de Ciencias, Universidad Nacional Autónoma de México, Ciudad de México, Ciudad de México, México 4 Laboratorio de Biología Evolutiva, Facultad de Ciencias, Universidad Autónoma del Estado de México, Toluca, Estado de México, México Corresponding Authors: Armando Sunny, Andrea González-Fernández Email address: [email protected], [email protected] Mexico is one of the most diverse countries that is losing a large amount of forest due to land use change, these data put Mexico in fourth place for global deforestation rate, therefore, Mexico occupies the first place in number of endangered species in the world with 665 endangered species. It is important to study amphibians because they are among the most threatened vertebrates on Earth and their populations are rapidly declining worldwide due primarily to the loss and degradation of their natural habitats. Pseudoeurycea robertsi is a micro-endemic and critically endangered Plethodontid salamander from the Nevado de Toluca Volcano and to date almost nothing is known about its natural history therefore, we survey fourteen sites of the Nevado de Toluca Volcano a mountain that is part of the Trans-Mexican Volcanic Belt, Mexico. -

Three New Species of Minute Salamanders (Plethodontidae: Thorius) from Oaxaca, Mexico
Biology of tiny animals: three new species of minute salamanders (Plethodontidae: Thorius) from Oaxaca, Mexico The Harvard community has made this article openly available. Please share how this access benefits you. Your story matters Citation Parra-Olea, Gabriela, Sean M. Rovito, Mario García-París, Jessica A. Maisano, David B. Wake, and James Hanken. 2016. “Biology of tiny animals: three new species of minute salamanders (Plethodontidae: Thorius) from Oaxaca, Mexico.” PeerJ 4 (1): e2694. doi:10.7717/ peerj.2694. http://dx.doi.org/10.7717/peerj.2694. Published Version doi:10.7717/peerj.2694 Citable link http://nrs.harvard.edu/urn-3:HUL.InstRepos:29626181 Terms of Use This article was downloaded from Harvard University’s DASH repository, and is made available under the terms and conditions applicable to Other Posted Material, as set forth at http:// nrs.harvard.edu/urn-3:HUL.InstRepos:dash.current.terms-of- use#LAA Biology of tiny animals: three new species of minute salamanders (Plethodontidae: Thorius) from Oaxaca, Mexico Gabriela Parra-Olea1, Sean M. Rovito2, Mario Garcı´a-Parı´s3, Jessica A. Maisano4,DavidB.Wake5 and James Hanken6 1 Instituto de Biologı´a, Universidad Nacional Auto´noma de Me´xico, Mexico City, Mexico 2 Unidad de Geno´mica Avanzada (Langebio), CINVESTAV, Irapuato, Guanajuato, Mexico 3 Departamento de Biodiversidad y Biologia Evolutiva, Museo Nacional de Ciencias Naturales (MNCN-CSIC), Madrid, Spain 4 Jackson School of Geosciences, University of Texas at Austin, Austin, Texas, United States 5 Department of Integrative Biology and Museum of Vertebrate Zoology, University of California, Berkeley, California, United States 6 Department of Organismic and Evolutionary Biology and Museum of Comparative Zoology, Harvard University, Cambridge, Massachusetts, United States ABSTRACT We describe three new species of minute salamanders, genus Thorius, from the Sierra Madre del Sur of Oaxaca, Mexico. -

Two New Species of Chiropterotriton (Caudata: Plethodontidae) from Central Veracruz, Mexico 1Mirna G
Offcial journal website: Amphibian & Reptile Conservation amphibian-reptile-conservation.org 12(2) [Special Section]: 37–54 (e167). urn:lsid:zoobank.org:pub:440CB3D6-450A-463B-B3D3-1CCBCBD8670E Two new species of Chiropterotriton (Caudata: Plethodontidae) from central Veracruz, Mexico 1Mirna G. García-Castillo, 2Ángel F. Soto-Pozos, 3J. Luis Aguilar-López, 4Eduardo Pineda, and 5Gabriela Parra-Olea 1,2,5Departamento de Zoología, Instituto de Biología, Universidad Nacional Autónoma de México, AP 70-153, Tercer Circuito Exterior s/n, Ciudad Universitaria, México, Distrito Federal, MÉXICO 3,4Red de Biología y Conservación de Vertebrados, Instituto de Ecología, A.C., Carretera Antigua a Coatepec No. 351, El Haya, CP. 91070, Xalapa, Veracruz, MÉXICO Abstract.—The lungless salamanders of the tribe Bolitoglossini show notable diversifcation in the Neotropics, and through the use of molecular tools and/or new discoveries, the total number of species continues to increase. Mexico is home to a great number of bolitoglossines primarily distributed along the eastern, central, and southern mountain ranges where the genus Chiropterotriton occurs. This group is relatively small, with 16 described species, but there remains a considerable number of undescribed species, suggested by the use of molecular tools in the lab more than a decade ago. Most of these undescribed species are found in the state of Veracruz, an area characterized by diverse topography and high salamander richness. Described herein are two new species of Chiropterotriton based on molecular and morphological data. Both new species were found in bromeliads in cloud forests of central Veracruz and do not correspond to any previously proposed species. Phylogenetic reconstructions included two mitochondrial fragments (L2 and COI) and identifed are two primary assemblages corresponding to northern and southern species distributions, concordant with previous studies. -

Diversification and Biogeographical History of Neotropical Plethodontid Salamanders
UC Berkeley UC Berkeley Previously Published Works Title Diversification and biogeographical history of Neotropical plethodontid salamanders Permalink https://escholarship.org/uc/item/7sk1k6vf Journal Zoological Journal of the Linnean Society, 175(1) ISSN 0024-4082 Authors Rovito, SM Parra-Olea, G Recuero, E et al. Publication Date 2015-09-01 DOI 10.1111/zoj.12271 Peer reviewed eScholarship.org Powered by the California Digital Library University of California bs_bs_banner Zoological Journal of the Linnean Society, 2015, 175, 167–188. With 8 figures Diversification and biogeographical history of Neotropical plethodontid salamanders SEAN M. ROVITO1,2†, GABRIELA PARRA-OLEA2*, ERNESTO RECUERO3 and DAVID B. WAKE1,4 1Museum of Vertebrate Zoology, University of California, 3101 Valley Life Sciences Building, Berkeley, CA 94720-3160, USA 2Instituto de Biología, Universidad Nacional Autónoma de México, AP 70-153, Circuito Exterior, Ciudad Universitaria, México, DF, CP 04510, México 3Departamento de Ecología de la Biodiversidad, Instituto de Ecología, Universidad Nacional Autónoma de México, AP 70-275, Ciudad Universitaria, México, DF, 04510, Mexico 4Department of Integrative Biology, University of California, 3040 Valley Life Sciences Building, Berkeley, CA 94720-3140, USA Received 18 September 2014; revised 27 February 2015; accepted for publication 2 March 2015 The Neotropical bolitoglossine salamanders represent an impressive adaptive radiation, comprising roughly 40% of global salamander species diversity. Despite decades of morphological studies and molecular work, a robust multilocus phylogenetic hypothesis based on DNA sequence data is lacking for the group. We estimated species trees based on multilocus nuclear and mitochondrial data for all major lineages within the bolitoglossines, and used our new phylogenetic hypothesis to test traditional biogeographical scenarios and hypotheses of morphological evolution in the group. -
Plethodontidae Bolitoglossa
Molecular Phylogenetics and Evolution 149 (2020) 106841 Contents lists available at ScienceDirect Molecular Phylogenetics and Evolution journal homepage: www.elsevier.com/locate/ympev Vastly underestimated species richness of Amazonian salamanders T (Plethodontidae: Bolitoglossa) and implications about plethodontid diversification ⁎ Andrés F. Jaramilloa,b, , Ignacio De La Rivac, Juan M. Guayasamind,e, Juan C. Chaparrof,g, Giussepe Gagliardi-Urrutiaa,b,h,i, Roberto C. Gutiérrezj, Isabela Brckok, Carles Vilàl, Santiago Castroviejo-Fishera,b,m a Pos-Graduação em Ecologia e Evolução da Biodiversidade, Pontifícia Universidade Católica do Rio Grande do Sul (PUCRS), Brazil b Laboratorio de Sistemática de Vertebrados, Pontifícia Universidade Católica do Rio Grande do Sul (PUCRS), Brazil c Museo Nacional de Ciencias Naturales (MNCN-CSIC), Spain d Laboratorio de Biología Evolutiva, Instituto BIOSFERA-USFQ, Colegio de Ciencias Biológicas y Ambientales COCIBA, Universidad San Francisco de Quito (USFQ), Ecuador e University of North Carolina at Chapel Hill, Department of Biology, USA f Museo de Biodiversidad del Perú (MUBI), Peru g Museo de Historia Natural de la Universidad Nacional de San Antonio Abad del Cusco, Peru h Peruvian Center for Biodiversity and Conservation (PCB&C), Peru i Dirección de Investigación en Diversidad Biológica Terrestre Amazónica, Instituto de Investigaciones de la Amazonía Peruana (IIAP), Peru j Museo de Historia Natural de la Universidad Nacional de San Agustín de Arequipa (MUSA), Peru k Laboratório de Biologia Molecular, Instituto de Ciências Biológicas, Universidade Federal do Pará (UFPA), Brazil l Estación Biológica de Doñana (EBD-CSIC), Spain m Department of Herpetology, American Museum of Natural History, USA ARTICLE INFO ABSTRACT Keywords: We present data showing that the number of salamander species in Amazonia is vastly underestimated. -
A New Maximum Body Size Record for the Berry Cave Salamander
A peer-reviewed open-access journal Subterranean Biology 28:A new29–38 maximum (2018) body size record for the Berry Cave Salamander... 29 doi: 10.3897/subtbiol.28.30506 SHORT COMMUNICATION Subterranean Published by http://subtbiol.pensoft.net The International Society Biology for Subterranean Biology A new maximum body size record for the Berry Cave Salamander (Gyrinophilus gulolineatus) and genus Gyrinophilus (Caudata, Plethodontidae) with a comment on body size in plethodontid salamanders Nicholas S. Gladstone1, Evin T. Carter2, K. Denise Kendall Niemiller3, Lindsey E. Hayter4, Matthew L. Niemiller3 1 Department of Earth and Planetary Sciences, University of Tennessee, Knoxville, Tennessee 37916, USA 2 Department of Ecology and Evolutionary Biology, University of Tennessee, Knoxville, Tennessee 37916, USA 3 Department of Biological Sciences, The University of Alabama in Huntsville, Huntsville, Alabama 35899, USA 4 Admiral Veterinary Hospital, 204 North Watt Road, Knoxville, Tennessee 37934, USA Corresponding author: Matthew L. Niemiller ([email protected]) Academic editor: O. Moldovan | Received 12 October 2018 | Accepted 23 October 2018 | Published 16 November 2018 http://zoobank.org/2BCA9CF1-52C7-47B6-BDFB-25DCF9EA487A Citation: Gladstone NS, Carter ET, Niemiller KDK, Hayter LE, Niemiller ML (2018) A new maximum body size record for the Berry Cave Salamander (Gyrinophilus gulolineatus) and genus Gyrinophilus (Caudata, Plethodontidae) with a comment on body size in plethodontid salamanders. Subterranean Biology 28: 29–38. https://doi.org/10.3897/ subtbiol.28.30506 Abstract Lungless salamanders in the family Plethodontidae exhibit an impressive array of life history strategies and occur in a diversity of habitats, including caves. However, relationships between life history, habitat, and body size remain largely unresolved. -
A New Species of Isthmura (Caudata: Plethodontidae) from the Montane Cloud Forest of Central Veracruz, Mexico
Zootaxa 4277 (4): 573–582 ISSN 1175-5326 (print edition) http://www.mapress.com/j/zt/ Article ZOOTAXA Copyright © 2017 Magnolia Press ISSN 1175-5334 (online edition) https://doi.org/10.11646/zootaxa.4277.4.7 http://zoobank.org/urn:lsid:zoobank.org:pub:6E14549C-730F-40E8-839D-85FE1041D443 A new species of Isthmura (Caudata: Plethodontidae) from the montane cloud forest of central Veracruz, Mexico ADRIANA SANDOVAL-COMTE1, EDUARDO PINEDA1,4, SEAN M. ROVITO2 & RICARDO LURÍA-MANZANO3 1 Red de Biología y Conservación de Vertebrados. Instituto de Ecología, A. C. Carretera antigua a Coatepec No. 351, El Haya, Xalapa, Veracruz, México. E-mail: [email protected]; E-mail: [email protected] 2Unidad de Genómica Avanzada (LANGEBIO), CINVESTAV, Irapuato, Guanajuato, México. E-mail: [email protected] 3Laboratorio de Herpetología, Facultad de Ciencias Biológicas, Benemérita Universidad Autónoma de Puebla, C.P. 72570, Puebla, México. E-mail:[email protected]. 4Corresponding author. E-mail: [email protected] Abstract We describe a new plethodontid salamander species of the genus Isthmura, known only from one locality in the moun- tainous region of central Veracruz, Mexico. Like its congeners, Isthmura corrugata sp. nov. has a large and robust body, but it is easily distinguished from the other species in the genus by the absence of any spot or mark on the dorsum (except by dull reddish brown coloration on eyelids) and by extremely well-marked costal grooves separated by very pronounced costal folds. Based on an mtDNA phylogeny, the new species is most closely related to the geographically distant I. boneti and I. -

Pdf, 19.80 Mb
Chiropterotriton chico García-Castillo, Rovito, Wake, and Parra-Olea 2017. El Chico Salamander is a state endemic species known only from Parque Nacional El Chico, at elevations from 2,400 to 3,050 m in pine-oak forest (Frost 2019). In the original description the authors noted that “it is unlikely to occur more widely, because surrounding areas have been extensively surveyed” (García-Castillo et al. 2017: 502). Also noted in the original description is that “this species was previously considered as conspecifc with C. multidentatus and occurs in sympatry with C. dimidiatus and Aquiloeurycea cephalica. Likewise, Isthmura bellii has been collected very near sites where C. chico was once common…, but it is unknown if the two species occur in syntopy” (García-Castillo et al. 2017: 502). García-Castillo et al. (2017: 502–503) noted that this salamander was “incredibly abundant” when one of the authors of this paper (David B. Wake) visited the park in August of 1971, but that by the mid-1970s it had become “uncommon and then rare.” These authors also indicated (p. 503) that the decline of C. chico apparently is not related to habitat loss or disturbance. They concluded that this salamander should be judged to be Critically Endangered, based on the IUCN criteria B1 ab(v). Finally, García-Castillo et al. (2017: 503) noted that “Hidalgo includes an unusual region where two of the main mountain complexes of Mexico meet: the Trans-Mexican Volcanic Belt (TMVB) and Sierra Madre Oriental (SMO). These ranges are known to have a high degree of topographic, geologic and climatic variability [that] has promoted a high biodiversity…This is especially true for the herpetofauna from Hidalgo…which is the state with the highest number of species of Chiropterotriton (6), representing 37.5% of the described species of this genus.” Photo by Sean Rovito, courtesy of Mirna G.