The Origin of Cultivated Citrus As Inferred from Internal Transcribed Spacer and Chloroplast DNA Sequence and Amplified Fragment Length Polymorphism Fingerprints
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Known Host Plants of Huanglongbing (HLB) and Asian Citrus Psyllid
Known Host Plants of Huanglongbing (HLB) and Asian Citrus Psyllid Diaphorina Liberibacter citri Plant Name asiaticus Citrus Huanglongbing Psyllid Aegle marmelos (L.) Corr. Serr.: bael, Bengal quince, golden apple, bela, milva X Aeglopsis chevalieri Swingle: Chevalier’s aeglopsis X X Afraegle gabonensis (Swingle) Engl.: Gabon powder-flask X Afraegle paniculata (Schum.) Engl.: Nigerian powder- flask X Atalantia missionis (Wall. ex Wight) Oliv.: see Pamburus missionis X X Atalantia monophylla (L.) Corr.: Indian atalantia X Balsamocitrus dawei Stapf: Uganda powder- flask X X Burkillanthus malaccensis (Ridl.) Swingle: Malay ghost-lime X Calodendrum capense Thunb.: Cape chestnut X × Citroncirus webberi J. Ingram & H. E. Moore: citrange X Citropsis gilletiana Swingle & M. Kellerman: Gillet’s cherry-orange X Citropsis schweinfurthii (Engl.) Swingle & Kellerm.: African cherry- orange X Citrus amblycarpa (Hassk.) Ochse: djerook leemo, djeruk-limau X Citrus aurantiifolia (Christm.) Swingle: lime, Key lime, Persian lime, lima, limón agrio, limón ceutí, lima mejicana, limero X X Citrus aurantium L.: sour orange, Seville orange, bigarde, marmalade orange, naranja agria, naranja amarga X Citrus depressa Hayata: shiikuwasha, shekwasha, sequasse X Citrus grandis (L.) Osbeck: see Citrus maxima X Citrus hassaku hort. ex Tanaka: hassaku orange X Citrus hystrix DC.: Mauritius papeda, Kaffir lime X X Citrus ichangensis Swingle: Ichang papeda X Citrus jambhiri Lushington: rough lemon, jambhiri-orange, limón rugoso, rugoso X X Citrus junos Sieb. ex Tanaka: xiang -

Characteristic Volatile Fingerprints and Odor Activity Values in Different
molecules Article Characteristic Volatile Fingerprints and Odor Activity Values in Different Citrus-Tea by HS-GC-IMS and HS-SPME-GC-MS Heting Qi 1,2,3 , Shenghua Ding 1,2,3, Zhaoping Pan 1,2,3, Xiang Li 1,2,3 and Fuhua Fu 1,2,3,* 1 Longping Branch Graduate School, Hunan University, Changsha 410125, China; [email protected] (H.Q.); [email protected] (S.D.); [email protected] (Z.P.); [email protected] (X.L.) 2 Provincial Key Laboratory for Fruits and Vegetables Storage Processing and Quality Safety, Agricultural Product Processing Institute, Hunan Academy of Agricultural Sciences, Changsha 410125, China 3 Hunan Province International Joint Lab on Fruits & Vegetables Processing, Quality and Safety, Changsha 410125, China * Correspondence: [email protected]; Tel.: +86-0731-82873369 Received: 24 November 2020; Accepted: 16 December 2020; Published: 19 December 2020 Abstract: Citrus tea is an emerging tea drink produced from tea and the pericarp of citrus, which consumers have increasingly favored due to its potential health effects and unique flavor. This study aimed to simultaneously combine the characteristic volatile fingerprints with the odor activity values (OAVs) of different citrus teas for the first time by headspace gas chromatography-ion mobility spectrometry (HS-GC-IMS) and headspace solid-phase microextraction-gas chromatography-mass spectrometry (HS-SPME-GC-MS). Results showed that the establishment of a citrus tea flavor fingerprint based on HS-GC-IMS data can provide an effective means for the rapid identification and traceability of different citrus varieties. Moreover, 68 volatile compounds (OAV > 1) were identified by HS-SPME-GC-MS, which reflected the contribution of aroma compounds to the characteristic flavor of samples. -

Organic Acids in the Juice of Acid Lemon and Japanese Acid Citrus by Gas Chromatography
九州大学学術情報リポジトリ Kyushu University Institutional Repository Organic Acids in the Juice of Acid Lemon and Japanese Acid Citrus by Gas Chromatography Widodo, Soesiladi E. Fruit Science Laboratory, Faculty of Agriculture, Kyushu University Shiraishi, Mikio Fruit Science Laboratory, Faculty of Agriculture, Kyushu University Shiraishi, Shinichi Fruit Science Laboratory, Faculty of Agriculture, Kyushu University https://doi.org/10.5109/24091 出版情報:九州大学大学院農学研究院紀要. 40 (1/2), pp.39-44, 1995-12. 九州大学農学部 バージョン: 権利関係: ,J. Fat. Agr., Kyushu IJniv., 40 (l-a), 39-44 (1995) Organic Acids in the Juice of Acid Lemon and Japanese Acid Citrus by Gas Chromatography Soesiladi E. Widodo, Mikio Shiraishi and Shinichi Shiraishi Fruit. Science Laboratory, Faculty of Agriculture, Kyushu University, Fukuoka 81 l-23, Japan (RWC~i/‘~?C~ C/1/?1P 15, 199<5) Acetate, glycolate, butyratc, oxalate, malonate, succinate, furnaratp, glyoxylate, malate, tattarate, cis-aconitatc and citrate were detected in the juice of Hanayu (Ci7tnt.s /ttrr/c~jrr Hart. ex Shirai), Daidai (C:. tr/i,rnt/tGt /II Linn. var. Cynthifera Y. Tanaka), Kabosu (6’. .sp/~rc:r~oc~r ~IXI Hart,. cx Tanaka), ‘Lisbon lemon (C.limon Burm. f. Lisbon) and Yuzu (C:.,jrr/ros Sieb. ex Tanaka) with compositions and contcnt,s varied according t,o sampling years and species. Citrate and rnalat,e were predominant, accounting for more than 90% and 3-9% of the total detected acids, respect.ively. The other acids presented in tracts, accounting t.ol.ally for roughly less than 0.5%. INTRODUCTION A number of chromatographic methods have been employed for determining organic acids (OAs) in citrus extracts. -
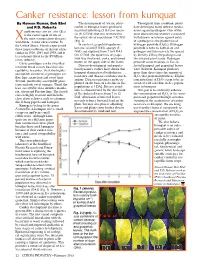
Canker Resistance: Lesson from Kumquat by Naveen Kumar, Bob Ebel the Development of Asiatic Citrus Throughout Their Evolution, Plants and P.D
Canker resistance: lesson from kumquat By Naveen Kumar, Bob Ebel The development of Asiatic citrus Throughout their evolution, plants and P.D. Roberts canker in kumquat leaves produced have developed many defense mecha- anthomonas citri pv. citri (Xcc) localized yellowing (5 DAI) or necro- nisms against pathogens. One of the is the causal agent of one of sis (9-12 DAI) that was restricted to most characteristic features associated the most serious citrus diseases the actual site of inoculation 7-12 DAI with disease resistance against entry X (Fig. 2). of a pathogen is the production of worldwide, Asiatic citrus canker. In the United States, Florida experienced In contrast, grapefruit epidermis hydrogen peroxide (H2O2). Hydrogen three major outbreaks of Asiatic citrus became raised (5 DAI), spongy (5 peroxide is toxic to both plant and canker in 1910, 1984 and 1995, and it DAI) and ruptured from 7 to 8 DAI. pathogen and thus restricts the spread is a constant threat to the $9 billion On 12 DAI, the epidermis of grape- by directly killing the pathogen and citrus industry. fruit was thickened, corky, and turned the infected plant tissue. Hydrogen Citrus genotypes can be classified brown on the upper side of the leaves. peroxide concentrations in Xcc-in- into four broad classes based on sus- Disease development and popula- fected kumquat and grapefruit leaves ceptibility to canker. First, the highly- tion dynamics studies have shown that were different. Kumquat produces susceptible commercial genotypes are kumquat demonstrated both disease more than three times the amount of Key lime, grapefruit and sweet lime. -

Freeze Response of Citrus and Citrus- Speeds (Nisbitt Et Al., 2000)
HORTSCIENCE 49(8):1010–1016. 2014. and tree and grove size (Bourgeois et al., 1990; Ebel et al., 2005). Protection using microsprinklers is compromised by high wind Freeze Response of Citrus and Citrus- speeds (Nisbitt et al., 2000). Developing more cold-tolerant citrus varieties through breeding related Genotypes in a Florida Field and selection has long been considered the most effective long-term solution (Grosser Planting et al., 2000; Yelenosky, 1985). Citrus and Citrus relatives are members Sharon Inch, Ed Stover1, and Randall Driggers of the family Rutaceae. The subtribe Citrinae U.S. Horticultural Research Laboratory, U.S. Department of Agriculture, is composed of Citrus (mandarins, oranges, Agricultural Research Service, 2001 South Rock Road, Fort Pierce, FL pummelos, grapefruits, papedas, limes, lem- ons, citrons, and sour oranges); Poncirus 34945 (deciduous trifoliate oranges); Fortunella Richard F. Lee (kumquats); Microcitrus and Eremocitrus (both Australian natives); and Clymenia National Clonal Germplasm Repository for Citrus and Dates, U.S. (Penjor et al., 2013). There is considerable Department of Agriculture, Agricultural Research Service, 1060 Martin morphological and ecological variation within Luther King Boulevard, Riverside, CA 92521 this group. With Citrus, cold-hardiness ranges from cold-tolerant to cold-sensitive (Soost and Additional index words. Aurantioideae, citrus breeding, cold-sensitive, defoliation, dieback, Roose, 1996). Poncirus and Fortunella are frost damage, Rutaceae, Toddalioideae considered the most cold-tolerant genera that Abstract. A test population consisting of progenies of 92 seed-source genotypes (hereafter are cross-compatible with Citrus. Poncirus called ‘‘parent genotypes’’) of Citrus and Citrus relatives in the field in east–central trifoliata reportedly can withstand tempera- Florida was assessed after natural freeze events in the winters of 2010 and 2011. -

PRUNING GUIDELINES Tools: Loppers, Saw, Clippers 10% Bleach and Water Solution
Varieties - choose one that you will want to eat often, as you will have them much of the year (Four Winds Citrus Variety Chart link in your Resources list) - certain citrus mature earlier than others (see early ripening handout) - Unique varieties: - Blood orange: red flesh is antioxidant rich. Often sweeter than other oranges - Yuzu: very little, but very flavorful juice used by chefs. Believed to be a cross between a sour mandarin and ichang papeda - Keiffir lime: regular lime with bumpy skin. Attractive tree with segmented leaves that are extremely fragrant and prized by chefs. - Buddhas hand: not much juice, but very fragrant pith and rind. Odd shaped and often used as an ornamental - Australian finger lime: oblong green lime with many small lime-flavored orbs inside. Called "citrus caviar" Standard varieties of citrus trees often grow to a height of 20 to 30 feet and the canopy -- or width of a tree -- can spread to 18 to 30 feet depending on the variety. Dwarf citrus trees are significantly shorter and narrower, which provides greater flexibility in planting location. Most varieties top out at 8 feet in height with a proportionally smaller canopy. Despite the differences in height and width, regular, semi-dwarf and dwarf citrus varieties produce the same size fruit. -First off DO NOT PRUNE! Those damaged leaves can actually provide protection for the plant until the air warms up. The plant needs to rally and recover. Pruning might just put it over the edge. Sometimes the plants must remain with that ‘raggedy’ appearance until as late as June and in some cases a full year. -

Citrus from Seed?
Which citrus fruits will come true to type Orogrande, Tomatera, Fina, Nour, Hernandina, Clementard.) from seed? Ellendale Tom McClendon writes in Hardy Citrus Encore for the South East: Fortune Fremont (50% monoembryonic) “Most common citrus such as oranges, Temple grapefruit, lemons and most mandarins Ugli Umatilla are polyembryonic and will come true to Wilking type. Because most citrus have this trait, Highly polyembryonic citrus types : will mostly hybridization can be very difficult to produce nucellar polyembryonic seeds that will grow true to type. achieve…. This unique characteristic Citrus × aurantiifolia Mexican lime (Key lime, West allows amateurs to grow citrus from seed, Indian lime) something you can’t do with, say, Citrus × insitorum (×Citroncirus webberii) Citranges, such as Rusk, Troyer etc. apples.” [12*] Citrus × jambhiri ‘Rough lemon’, ‘Rangpur’ lime, ‘Otaheite’ lime Monoembryonic (don’t come true) Citrus × limettioides Palestine lime (Indian sweet lime) Citrus × microcarpa ‘Calamondin’ Meyer Lemon Citrus × paradisi Grapefruit (Marsh, Star Ruby, Nagami Kumquat Redblush, Chironja, Smooth Flat Seville) Marumi Kumquat Citrus × sinensis Sweet oranges (Blonde, navel and Pummelos blood oranges) Temple Tangor Citrus amblycarpa 'Nasnaran' mandarin Clementine Mandarin Citrus depressa ‘Shekwasha’ mandarin Citrus karna ‘Karna’, ‘Khatta’ Poncirus Trifoliata Citrus kinokuni ‘Kishu mandarin’ Citrus lycopersicaeformis ‘Kokni’ or ‘Monkey mandarin’ Polyembryonic (come true) Citrus macrophylla ‘Alemow’ Most Oranges Citrus reshni ‘Cleopatra’ mandarin Changshou Kumquat Citrus sunki (Citrus reticulata var. austera) Sour mandarin Meiwa Kumquat (mostly polyembryonic) Citrus trifoliata (Poncirus trifoliata) Trifoliate orange Most Satsumas and Tangerines The following mandarin varieties are polyembryonic: Most Lemons Dancy Most Limes Emperor Grapefruits Empress Tangelos Fairchild Kinnow Highly monoembryonic citrus types: Mediterranean (Avana, Tardivo di Ciaculli) Will produce zygotic monoembryonic seeds that will not Naartje come true to type. -
Holdings of the University of California Citrus Variety Collection 41
Holdings of the University of California Citrus Variety Collection Category Other identifiers CRC VI PI numbera Accession name or descriptionb numberc numberd Sourcee Datef 1. Citron and hybrid 0138-A Indian citron (ops) 539413 India 1912 0138-B Indian citron (ops) 539414 India 1912 0294 Ponderosa “lemon” (probable Citron ´ lemon hybrid) 409 539491 Fawcett’s #127, Florida collection 1914 0648 Orange-citron-hybrid 539238 Mr. Flippen, between Fullerton and Placentia CA 1915 0661 Indian sour citron (ops) (Zamburi) 31981 USDA, Chico Garden 1915 1795 Corsican citron 539415 W.T. Swingle, USDA 1924 2456 Citron or citron hybrid 539416 From CPB 1930 (Came in as Djerok which is Dutch word for “citrus” 2847 Yemen citron 105957 Bureau of Plant Introduction 3055 Bengal citron (ops) (citron hybrid?) 539417 Ed Pollock, NSW, Australia 1954 3174 Unnamed citron 230626 H. Chapot, Rabat, Morocco 1955 3190 Dabbe (ops) 539418 H. Chapot, Rabat, Morocco 1959 3241 Citrus megaloxycarpa (ops) (Bor-tenga) (hybrid) 539446 Fruit Research Station, Burnihat Assam, India 1957 3487 Kulu “lemon” (ops) 539207 A.G. Norman, Botanical Garden, Ann Arbor MI 1963 3518 Citron of Commerce (ops) 539419 John Carpenter, USDCS, Indio CA 1966 3519 Citron of Commerce (ops) 539420 John Carpenter, USDCS, Indio CA 1966 3520 Corsican citron (ops) 539421 John Carpenter, USDCS, Indio CA 1966 3521 Corsican citron (ops) 539422 John Carpenter, USDCS, Indio CA 1966 3522 Diamante citron (ops) 539423 John Carpenter, USDCS, Indio CA 1966 3523 Diamante citron (ops) 539424 John Carpenter, USDCS, Indio -

Chemical Variability of Peel and Leaf Essential Oils in the Citrus Subgenus Papeda (Swingle) and Few Relatives
plants Article Chemical Variability of Peel and Leaf Essential Oils in the Citrus Subgenus Papeda (Swingle) and Few Relatives Clémentine Baccati 1, Marc Gibernau 1, Mathieu Paoli 1 , Patrick Ollitrault 2,3 ,Félix Tomi 1,* and François Luro 2 1 Laboratoire Sciences Pour l’Environnement, Equipe Chimie et Biomasse, Université de Corse—CNRS, UMR 6134 SPE, Route des Sanguinaires, 20000 Ajaccio, France; [email protected] (C.B.); [email protected] (M.G.); [email protected] (M.P.) 2 UMR AGAP Institut, Université Montpellier, CIRAD, INRAE, Institut Agro, 20230 San Giuliano, France; [email protected] (P.O.); [email protected] (F.L.) 3 CIRAD, UMR AGAP, 20230 San Giuliano, France * Correspondence: [email protected]; Tel.: +33-495-52-4122 Abstract: The Papeda Citrus subgenus includes several species belonging to two genetically distinct groups, containing mostly little-exploited wild forms of citrus. However, little is known about the potentially large and novel aromatic diversity contained in these wild citruses. In this study, we characterized and compared the essential oils obtained from peels and leaves from representatives of both Papeda groups, and three related hybrids. Using a combination of GC, GC-MS, and 13C-NMR spectrometry, we identified a total of 60 compounds in peel oils (PO), and 76 compounds in leaf oils (LO). Limonene was the major component in almost all citrus PO, except for C. micrantha and C. hystrix, where β-pinene dominated (around 35%). LO composition was more variable, with different Citation: Baccati, C.; Gibernau, M.; major compounds among almost all samples, except for two citrus pairs: C. -

Nobu Miami Beverage Drinks Menu
NOBU'S SAKE SELECTION The Hokusetsu Brewery on Sado Island in the Sea of Japan has been operated by the Hazu family since 1871. The name "Hokusetsu", or “Northern Snow”, was chosen to reflect the ideal sake brewing conditions on the island during the coldest days of winter. Chef Nobu first experienced Hokusetsu sake when his Japanese rock-musician friend brought a bottle to the original Matsuhisa restaurant in Beverly Hills. Impressed by the quality, Nobu obtained exclusive rights to sell Hokusetsu sake in the United States. JUNMAI DAIGINJO TK40 GENSHU ENSHINBUNRI 'HIKARI' The rare hybrid rice grain, Koshitanrei, is polished to 40% of its original size to produce this premium sake. Elegant floral flavors of Orange Blossom, Elderflower, and White Tea are found in this powerful Genshu, or undiluted sake. Bottle 24 oz $680 DAIGINJO YK35 SHIZUKU The most sought-after rice grain, Yamada Nishiki, is polished to 35% of its original size to produce this premium sake. The sake is extracted through a slow drip process that highlights delicate flavors of Lychee, Honeydew, and Pear. Fruit forward and amazingly smooth. Bottle 24 oz $560 DAIGINJO YK35 SHIZUKU JUKUSEI KOSHU This is the YK35 Shizuku, aged for three years. The aging process has added structure to the delicate Nashi Pear flavor becoming highly complex with a deep richness not found in young sake. This is made in extremely limited quantities. Bottle 60 oz $3500 NOBU SAKE SELECTION GINJO NIGORI Unfiltered, dry and creamy Glass Bottle 16 oz $12 $64 Pepino Light-bodied crisp and dry with a -

Citrus Rootstocks: Their Characters and Reactions
CITRUS ROOTSTOCKS: THEIR CHARACTERS AND REACTIONS (an unpublished manuscript) ca. 1986 By W. P. BITTERS (1915 – 2006) Editor, digital version: Marty Nemeth, Reference Librarian, UC Riverside Science Library, retired Subject matter experts, digital version: Dr. Tracy Kahn, Curator, UC Citrus Variety Collection Dr. Robert Krueger, Curator, USDA-ARS National Clonal Germplasm Repository for Citrus & Dates Toni Siebert, Assistant Curator, UC Citrus Variety Collection ca. 1955 ca. 1970 IN MEMORIUM Willard P. Bitters Professor of Horticulture, Emeritus Riverside 1915-2006 Born in Eau Claire, Wisconsin, in June, 1915, Dr. Willard “Bill” Bitters earned his bachelor’s degree in biology from St. Norbert College and his master’s degree and Ph.D. from the University of Wisconsin. After earning his doctorate, he first worked as the superintendent of the Valley Research Farm of the University of Arizona in Yuma, and joined the Citrus Experiment Station, in Riverside in 1946 as a Horticulturist. In 1961, Dr. Bitters became a Professor in the newly established University of California-Riverside. His initial assignment was to work on horticultural aspects of tristeza, a serious vector-transmitted virus disease which threatened to destroy California citrus orchards. Tristeza was already in California and spreading in 1946. At that time most citrus trees in California were grafted on a rootstock that was known to be susceptible to tristeza. Dr. Bill Bitters was responsible for screening of over 500 cultivars to determine which rootstock-scion combinations were resistant to this disease and yet possessed suitable horticultural characteristics. Of the 500 screened, most were susceptible, but several successful ones were selected and released to the industry. -

CITRUS BUDWOOD Annual Report 2017-2018
CITRUS BUDWOOD Annual Report 2017-2018 Citrus Nurseries affected by Hurricane Irma, September 2017 Florida Department of Agriculture and Consumer Services Our Vision The Bureau of Citrus Budwood Registration will be diligent in providing high yielding, pathogen tested, quality budlines that will positively impact the productivity and prosperity of our citrus industry. Our Mission The Bureau of Citrus Budwood Registration administers a program to assist growers and nurserymen in producing citrus nursery trees that are believed to be horticulturally true to varietal type, productive, and free from certain recognizable bud-transmissible diseases detrimental to fruit production and tree longevity. Annual Report 2018 July 1, 2017 – June 30, 2018 Bureau of Citrus Budwood Registration Ben Rosson, Chief This is the 64th year of the Citrus Budwood Registration Program which began in Florida in 1953. Citrus budwood registration and certification programs are vital to having a healthy commercial citrus industry. Clean stock emerging from certification programs is the best way to avoid costly disease catastrophes in young plantings and their spread to older groves. Certification programs also restrict or prevent pathogens from quickly spreading within growing areas. Regulatory endeavors have better prospects of containing or eradicating new disease outbreaks if certification programs are in place to control germplasm movement. Budwood registration has the added benefit in allowing true-to-type budlines to be propagated. The selection of high quality cultivars for clonal propagation gives growers uniform plantings of high quality trees. The original mother stock selected for inclusion in the Florida budwood program is horticulturally evaluated for superior performance, either by researchers, growers or bureau staff.