The Role of YB1 in Renal Cell Carcinoma Cell Adhesion
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Biological Pathways and in Vivo Antitumor Activity Induced by Atiprimod in Myeloma
Leukemia (2007) 21, 2519–2526 & 2007 Nature Publishing Group All rights reserved 0887-6924/07 $30.00 www.nature.com/leu ORIGINAL ARTICLE Biological pathways and in vivo antitumor activity induced by Atiprimod in myeloma P Neri1,2,3, P Tassone1,2,3, M Shammas1, H Yasui2, E Schipani4, RB Batchu1, S Blotta1,2,3, R Prabhala1, L Catley2, M Hamasaki2, T Hideshima2, D Chauhan2, GS Jacob5, D Picker5, S Venuta3, KC Anderson2 and NC Munshi1,2 1Jerome Lipper Multiple Myeloma Center, Department of Adult Oncology, Dana-Farber Cancer Institute, Harvard Medical School, Boston, MA, USA; 2Boston VA Healthcare System, Department of Medicine, Harvard Medical School, MA, USA; 3Department of Experimental and Clinical Medicine, University of ‘Magna Græcia’ and Cancer Center, Catanzaro, Italy; 4Endocrine Unit, Massachusetts General Hospital, Boston, MA, USA and 5Callisto Pharmaceuticals Inc., New York, NY, USA Atiprimod (Atip) is a novel oral agent with anti-inflammatory tion.7,8 Atip inhibits the inflammatory response and preserves properties. Although its in vitro activity and effects on signaling bone integrity in murine models of rheumatoid arthritis (RA),9–12 in multiple myeloma (MM) have been previously reported, here targets macrophages, inhibits phospholipase A and C in rat we investigated its molecular and in vivo effects in MM. Gene 13,14 expression analysis of MM cells identified downregulation of alveolar macrophages and exhibits antiproliferative and 15–17 genes involved in adhesion, cell-signaling, cell cycle and bone antiangiogenic activities in human cancer models. Impor- morphogenetic protein (BMP) pathways and upregulation of tantly, we have previously reported that Atip inhibits MM cell genes implicated in apoptosis and bone development, follow- growth, induces caspase-mediated apoptosis, blocks the phos- ing Atip treatment. -

Supplementary Table 1: Adhesion Genes Data Set
Supplementary Table 1: Adhesion genes data set PROBE Entrez Gene ID Celera Gene ID Gene_Symbol Gene_Name 160832 1 hCG201364.3 A1BG alpha-1-B glycoprotein 223658 1 hCG201364.3 A1BG alpha-1-B glycoprotein 212988 102 hCG40040.3 ADAM10 ADAM metallopeptidase domain 10 133411 4185 hCG28232.2 ADAM11 ADAM metallopeptidase domain 11 110695 8038 hCG40937.4 ADAM12 ADAM metallopeptidase domain 12 (meltrin alpha) 195222 8038 hCG40937.4 ADAM12 ADAM metallopeptidase domain 12 (meltrin alpha) 165344 8751 hCG20021.3 ADAM15 ADAM metallopeptidase domain 15 (metargidin) 189065 6868 null ADAM17 ADAM metallopeptidase domain 17 (tumor necrosis factor, alpha, converting enzyme) 108119 8728 hCG15398.4 ADAM19 ADAM metallopeptidase domain 19 (meltrin beta) 117763 8748 hCG20675.3 ADAM20 ADAM metallopeptidase domain 20 126448 8747 hCG1785634.2 ADAM21 ADAM metallopeptidase domain 21 208981 8747 hCG1785634.2|hCG2042897 ADAM21 ADAM metallopeptidase domain 21 180903 53616 hCG17212.4 ADAM22 ADAM metallopeptidase domain 22 177272 8745 hCG1811623.1 ADAM23 ADAM metallopeptidase domain 23 102384 10863 hCG1818505.1 ADAM28 ADAM metallopeptidase domain 28 119968 11086 hCG1786734.2 ADAM29 ADAM metallopeptidase domain 29 205542 11085 hCG1997196.1 ADAM30 ADAM metallopeptidase domain 30 148417 80332 hCG39255.4 ADAM33 ADAM metallopeptidase domain 33 140492 8756 hCG1789002.2 ADAM7 ADAM metallopeptidase domain 7 122603 101 hCG1816947.1 ADAM8 ADAM metallopeptidase domain 8 183965 8754 hCG1996391 ADAM9 ADAM metallopeptidase domain 9 (meltrin gamma) 129974 27299 hCG15447.3 ADAMDEC1 ADAM-like, -

Anti-Integrin Beta 8 Antibody (ARG56852)
Product datasheet [email protected] ARG56852 Package: 100 μl anti-Integrin beta 8 antibody Store at: -20°C Summary Product Description Rabbit Polyclonal antibody recognizes Integrin beta 8 Tested Reactivity Hu, Ms, Rat Tested Application WB Host Rabbit Clonality Polyclonal Isotype IgG Target Name Integrin beta 8 Antigen Species Human Immunogen Recombinant protein of Human Integrin beta 8. Conjugation Un-conjugated Alternate Names Integrin beta-8 Application Instructions Application table Application Dilution WB 1:500 - 1:2000 Application Note * The dilutions indicate recommended starting dilutions and the optimal dilutions or concentrations should be determined by the scientist. Positive Control 293T Calculated Mw 86 kDa Properties Form Liquid Purification Affinity purification. Buffer PBS (pH 7.3), 0.02% Sodium azide and 50% Glycerol. Preservative 0.02% Sodium azide Stabilizer 50% Glycerol Storage instruction For continuous use, store undiluted antibody at 2-8°C for up to a week. For long-term storage, aliquot and store at -20°C. Storage in frost free freezers is not recommended. Avoid repeated freeze/thaw cycles. Suggest spin the vial prior to opening. The antibody solution should be gently mixed before use. Note For laboratory research only, not for drug, diagnostic or other use. www.arigobio.com 1/2 Bioinformation Database links GeneID: 3696 Human Swiss-port # P26012 Human Gene Symbol ITGB8 Gene Full Name integrin, beta 8 Background This gene is a member of the integrin beta chain family and encodes a single-pass type I membrane protein with a VWFA domain and four cysteine-rich repeats. This protein noncovalently binds to an alpha subunit to form a heterodimeric integrin complex. -

ID AKI Vs Control Fold Change P Value Symbol Entrez Gene Name *In
ID AKI vs control P value Symbol Entrez Gene Name *In case of multiple probesets per gene, one with the highest fold change was selected. Fold Change 208083_s_at 7.88 0.000932 ITGB6 integrin, beta 6 202376_at 6.12 0.000518 SERPINA3 serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 3 1553575_at 5.62 0.0033 MT-ND6 NADH dehydrogenase, subunit 6 (complex I) 212768_s_at 5.50 0.000896 OLFM4 olfactomedin 4 206157_at 5.26 0.00177 PTX3 pentraxin 3, long 212531_at 4.26 0.00405 LCN2 lipocalin 2 215646_s_at 4.13 0.00408 VCAN versican 202018_s_at 4.12 0.0318 LTF lactotransferrin 203021_at 4.05 0.0129 SLPI secretory leukocyte peptidase inhibitor 222486_s_at 4.03 0.000329 ADAMTS1 ADAM metallopeptidase with thrombospondin type 1 motif, 1 1552439_s_at 3.82 0.000714 MEGF11 multiple EGF-like-domains 11 210602_s_at 3.74 0.000408 CDH6 cadherin 6, type 2, K-cadherin (fetal kidney) 229947_at 3.62 0.00843 PI15 peptidase inhibitor 15 204006_s_at 3.39 0.00241 FCGR3A Fc fragment of IgG, low affinity IIIa, receptor (CD16a) 202238_s_at 3.29 0.00492 NNMT nicotinamide N-methyltransferase 202917_s_at 3.20 0.00369 S100A8 S100 calcium binding protein A8 215223_s_at 3.17 0.000516 SOD2 superoxide dismutase 2, mitochondrial 204627_s_at 3.04 0.00619 ITGB3 integrin, beta 3 (platelet glycoprotein IIIa, antigen CD61) 223217_s_at 2.99 0.00397 NFKBIZ nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, zeta 231067_s_at 2.97 0.00681 AKAP12 A kinase (PRKA) anchor protein 12 224917_at 2.94 0.00256 VMP1/ mir-21likely ortholog -

Fibroblasts from the Human Skin Dermo-Hypodermal Junction Are
cells Article Fibroblasts from the Human Skin Dermo-Hypodermal Junction are Distinct from Dermal Papillary and Reticular Fibroblasts and from Mesenchymal Stem Cells and Exhibit a Specific Molecular Profile Related to Extracellular Matrix Organization and Modeling Valérie Haydont 1,*, Véronique Neiveyans 1, Philippe Perez 1, Élodie Busson 2, 2 1, 3,4,5,6, , Jean-Jacques Lataillade , Daniel Asselineau y and Nicolas O. Fortunel y * 1 Advanced Research, L’Oréal Research and Innovation, 93600 Aulnay-sous-Bois, France; [email protected] (V.N.); [email protected] (P.P.); [email protected] (D.A.) 2 Department of Medical and Surgical Assistance to the Armed Forces, French Forces Biomedical Research Institute (IRBA), 91223 CEDEX Brétigny sur Orge, France; [email protected] (É.B.); [email protected] (J.-J.L.) 3 Laboratoire de Génomique et Radiobiologie de la Kératinopoïèse, Institut de Biologie François Jacob, CEA/DRF/IRCM, 91000 Evry, France 4 INSERM U967, 92260 Fontenay-aux-Roses, France 5 Université Paris-Diderot, 75013 Paris 7, France 6 Université Paris-Saclay, 78140 Paris 11, France * Correspondence: [email protected] (V.H.); [email protected] (N.O.F.); Tel.: +33-1-48-68-96-00 (V.H.); +33-1-60-87-34-92 or +33-1-60-87-34-98 (N.O.F.) These authors contributed equally to the work. y Received: 15 December 2019; Accepted: 24 January 2020; Published: 5 February 2020 Abstract: Human skin dermis contains fibroblast subpopulations in which characterization is crucial due to their roles in extracellular matrix (ECM) biology. -

Transcriptome-Wide Analysis of CXCR5 Deficient Retinal Pigment
Article Transcriptome‐wide analysis of CXCR5 deficient retinal pigment epithelial (RPE) cells reveals molecular signature of RPE homeostasis Supplementary data Madhu Sudhana Saddala 1, Anton Lennikov 1, Anthony Mukwaya 2, and Hu Huang 1,* 1 Department of Ophthalmology, University of Missouri, Columbia, MO 65212, United States of America; [email protected] (M.S.S.); [email protected] (A.L.) 2 Department of Ophthalmology, Institute for Clinical and Experimental Medicine, Faculty of Health Sciences, Linköping University, Linköping, 58183 , Sweden; [email protected] * Correspondence: [email protected]; Tel: +1 573‐882‐9899 # Madhu Sudhana Saddala and Anton Lennikov have contributed equally to this work. Keywords: Age‐related macular degeneration; CXCR5; EMT, FoxO; Mitochondria; RNA‐Seq, Gene Ontology; KEGG; Retinal pigment epithelium Supplementary Figures Biomedicines 2020, 8, x; doi: FOR PEER REVIEW www.mdpi.com/journal/biomedicines Biomolecules 2020, 10, x FOR PEER REVIEW 2 of 13 Biomolecules 2020, 10, x FOR PEER REVIEW 3 of 13 Figure S1: Quantitative profiling of mouse RPE tissue between control and CXCR5 ko groups. Pearson correlation coefficient plot of the log2 ratio between two groups (A) before and (B) after normalization. Potential relationships or correlations amongst the different data attributes is to leverage a pair‐wise correlation matrix in control and CXCR5 knockout groups. Biomolecules 2020, 10, x FOR PEER REVIEW 4 of 13 Biomolecules 2020, 10, x FOR PEER REVIEW 5 of 13 Figure S2: Potential relationships or correlations amongst the different data attributes is to leverage a pair‐wise correlation matrix in control and CXCR5 knockout groups. (A) heatmap of total differentially expressed genes (B) The cluster of main heatmap represented twenty differentially expressed genes. -

PP1-Associated Signaling and − B/AP-1 Κ Inhibition of NF- Tolerance
Downloaded from http://www.jimmunol.org/ by guest on October 3, 2021 is online at: average * and − B/AP-1 κ The Journal of Immunology published online 26 February 2014 from submission to initial decision 4 weeks from acceptance to publication http://www.jimmunol.org/content/early/2014/02/26/jimmun ol.1301610 Identification of Two Forms of TNF Tolerance in Human Monocytes: Differential Inhibition of NF- PP1-Associated Signaling Johannes Günther, Nico Vogt, Katharina Hampel, Rolf Bikker, Sharon Page, Benjamin Müller, Judith Kandemir, Michael Kracht, Oliver Dittrich-Breiholz, René Huber and Korbinian Brand J Immunol Submit online. Every submission reviewed by practicing scientists ? is published twice each month by Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts http://jimmunol.org/subscription Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html http://www.jimmunol.org/content/suppl/2014/02/26/jimmunol.130161 0.DCSupplemental Information about subscribing to The JI No Triage! Fast Publication! Rapid Reviews! 30 days* Why • • • Material Permissions Email Alerts Subscription Supplementary The Journal of Immunology The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2014 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. This information is current as of October 3, 2021. Published February 26, 2014, doi:10.4049/jimmunol.1301610 The Journal of Immunology Identification of Two Forms of TNF Tolerance in Human Monocytes: Differential Inhibition of NF-kB/AP-1– and PP1-Associated Signaling Johannes Gunther,*€ ,1 Nico Vogt,*,1 Katharina Hampel,*,1 Rolf Bikker,* Sharon Page,* Benjamin Muller,*€ Judith Kandemir,* Michael Kracht,† Oliver Dittrich-Breiholz,‡ Rene´ Huber,* and Korbinian Brand* The molecular basis of TNF tolerance is poorly understood. -
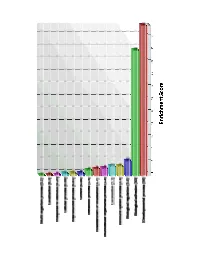
Supplementary Data
aabb d ccd a b c d ab a b c d Supplementary Figure S1: Gene ontology analysis of genes upregulated in pancreatic CAFs relative to control fibroblasts. A candidate list of 200 genes upregulated 2.8 -fold or more in pancreatic CAFs relative to control fibroblasts was sorted into representative biological function groups using the Partek Gene Ontology analysis. The high enrichment scores in the biological adhesion and developmental process groups indicate that the candidate gene list is enriched for genes involved in these processes. Supplementary Figure S2: Characterization of HPNE. Immunohistochemical staining reveals that HPNE cells express the mesenchymal marker, vimentin (a) and do not express the marker of activated fibroblasts, alpha-smooth muscle actin (b). HPNE cells lack expression of the cytokeratin 19 (c), an epithelial marker expressed in the pancreatic ductal epithelial cell line HPDE (d). Magnification: A: x20; B, C, D: x10. Supplementary Figure S3: Characterization of pancreatic CAFs. Cells within the CAF cultures have both a stellate and spindle-type morphology and lack expression of the epithelial cell marker cytokeratin 19 (a). They contain cytoplasmic lipid droplets (b) and express the mesenchymal cytoskeletal filament vimentin (c) as well as alpha-SMA (d), a marker of activated fibroblasts. Magnification: A, C, D: x10; B: x20. Figure is representative of all CAFs. Supplementary Figure S4: Immunohistochemical analysis of Smo expression in normal pancreatic tissue. Paraffin embedded sections were probed with an antibody against vimentin (a) or against Smo (b). Arrows indicate normal fibroblasts expressing vimentin but not Smo. Magnification: x20. 1 Supplementary Figure S5: Immunohistochemical analysis of Smo expression in whole tissue sections from pancreatic cancer tissue: Paraffin-embedded sections from primary pancreatic cancer tissue were probed with an antibody against alpha-SMA (a,c; low and high power images of the same section) to highlight the activated cancer associated fibroblasts. -
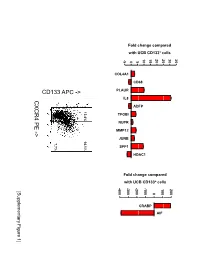
Supplementary Data
Fold change compared with UCB CD133+ cells 10 15 20 25 30 35 -5 0 5 COL4A1 CD68 PLAUR IL8 CXCR4 PE ADFP 13.4% 14.1% TFGBI PE NUPR - MMP12 > JUNB 7.7% SPP1 HDAC1 Fold change compared with UCB CD133+ cells -400 -300 -200 -100 100 200 [Supplementary Figure1] 0 CRABP AIF Rutella S et al. Supplementary Materials and Methods Real-time PCR Real-time quantitative PCR (qPCR) was performed on mRNA samples isolated from either endometrial cancer-derived CD133+ cells or umbilical cord blood CD133+ cells with the RNeasy plus Mini Kit (QIAGEN, Hilden, Germany), as already detailed. Quantitative PCR for a selected group of genes was performed on the iCycler iQ system (Bio-Rad, Hercules, CA). Primer sets were designed using the Beacon Design Software (Version 3) and the sequences available in the GeneBankTM data base. The specific oligonucleotide primer sequences are detailed in Supplementary Table 8. Complementary DNA (cDNA) was prepared starting from 1μg of total RNA using the iScrypt cDNA Synthesis Kit, which contains RNase H + MMLV reverse transcriptase random primers and 5x reaction mixture (Bio-Rad), according to the manufacturer’s instructions. Reactions were conducted in the PTC-0200 DNA Engine (MJ RESEARCH). Amplification was carried out in a total volume of 25μl containing 0.3μM of each specific primer, 12,5μl 2X SYBR Green Master Mix (100mM KCl, 40mM Tris-HCl, pH 8.4, 0.4mM of each dNTP, iTaq DNA polymerase, 6mM MgCl2, SYBR Green I, 20nM fluorescein, stabilizers; Bio-Rad) and 2μl of diluted cDNA. The PCR reactions were cycled starting with a 3-minute template denaturation step at 95°C followed by 40 cycles of 15 seconds at 95°C and 1 minute at 60°C. -
Alzheimer's Disease
bioRxiv preprint doi: https://doi.org/10.1101/189712; this version posted May 31, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Alzheimer’s disease: the large gene instability hypothesis Sourena Soheili-Nezhad, MD [email protected] Donders Centre for Cognitive Neuroimaging, Radboud University Medical Center, Nijmegen, Netherlands Abstract All drug trials of the Alzheimer’s disease (AD) have failed to slow the progression of dementia in phase III studies, and the most effective therapeutic strategy remains controversial due to the poorly understood disease mechanisms. For AD drug design, amyloid beta (Aβ) and its cascade have been the primary focus since decades ago, but mounting evidence indicates that the underpinning molecular pathways of AD are more complex than the classical reductionist models. Several genome-wide association studies (GWAS) have recently shed light on dark aspects of AD from a hypothesis-free perspective. Here, I use this novel insight to suggest that the amyloid cascade hypothesis may be a wrong model for AD therapeutic design. I review 23 novel genetic risk loci and show that, as a common theme, they code for receptor proteins and signal transducers of cell adhesion pathways, with clear implications in synaptic development, maintenance, and function. Contrary to the Aβ-based interpretation, but further reinforcing the unbiased genome-wide insight, the classical hallmark genes of AD including the amyloid precursor protein (APP), presenilins (PSEN), and APOE also take part in similar pathways of growth cone adhesion and contact-guidance during brain development. -

Cell Biological Function of Secretome of Adipose-Derived Stem Cells on Human Dermal Fibroblasts and Keratinocytes
Korean J. Microbiol. Biotechnol. Vol. 40, No. 2, 117–127 (2012) http://dx.doi.org/10.4014/kjmb.1204.04001 pISSN 1598-642X eISSN 2234-7305 Cell Biological Function of Secretome of Adipose-Derived Stem Cells on Human Dermal Fibroblasts and Keratinocytes Lee, Jae Seol1 and Jong-Hwan Lee1,2,3* 1Department of Biomaterial Control, Dong-Eui University, Busan 614-714, Korea 2Blue-Bio Regional Innovation Center, Dong-Eui University, Busan 614-714, Korea 3Department of Biotechnology and Bioengineering, Dong-Eui University, Busan 614-714, Korea Received : April 3, 2012 / Revised : June 7, 2012 / Accepted : June 9, 2012 The beneficial effects of adipose-derived stem cell conditioned media (ADSC-CM) for skin regeneration have previously been reported, despite the precise mechanism of how ADSC-CM promotes skin regeneration remaining unclear. ADSC-CM contains various secretomes and this may be a factor in it being a good resource for the treatment of skin conditions. It is also known that ADSC-CM produced in hypoxia conditions, in other words Advanced Adipose-Derived Stem cell Protein Extract (AAPE), has excellent skin regenerative properties. In this study, a human primary skin cell was devised to examine how AAPE affects human dermal fibroblast (HDF) and human keratinocyte (HK), which both play fundamental roles in skin regeneration. The promotion of collagen formation by HDFs was observed at 0.32 mg/ml of AAPE. AAPE treatment signifi- cantly stimulated stress fiber formation. DNA gene chips demonstrated that AAPE in HKs (p<0.05) affected the expression of 133 identifiable transcripts, which were associated with cell proliferation, migration, cell adhesion, and response to wounding. -

Tropism-Modified AAV Vectors Overcome Barriers to Successful Cutaneous Therapy
Vector engineering and delivery © The American Society of Gene & Cell Therapy original article original article Capsid-modified AAV for Cutaneous Gene Transfer © The American Society of Gene & Cell Therapy Tropism-modified AAV Vectors Overcome Barriers to Successful Cutaneous Therapy Jessica Sallach1,2, Giovanni Di Pasquale3, Fernando Larcher4, Nadine Niehoff1,5, Matthias Rübsam1,5, Anke Huber1,2, Jay Chiorini3, David Almarza4,10, Sabine A Eming1,5,6, Hikmet Ulus7, Stephen Nishimura8, Ulrich T Hacker9, Michael Hallek1,2, Carien M Niessen1,5,6 and Hildegard Büning1,2 1Center for Molecular Medicine Cologne (CMMC), University of Cologne, Cologne, Germany; 2Department I for Internal Medicine, University Hospital Cologne, Cologne, Germany; 3Molecular Physiology and Therapeutics Branch, National Institute of Dental and Craniofacial Research, National Insti- tutes of Health, Bethesda, Maryland, USA; 4Centro de Investigacion Energetica Medioambiental y Tecnologica (CIEMAT) Madrid, Center for Biomedical Research on Rare Diseases (CIBERER-U714) and Department of Bioengineering. Universidad Carlos III de Madrid (UC3M), Madrid, Spain; 5Depart- ment of Dermatology, University Hospital of Cologne, Cologne, Germany; 6Cologne Excellence Center on Cellular Stress Responses in Aging-Associated Diseases (CECAD), Cologne, Germany; 7Children’s Medical Surgery, Cologne, Germany; 8Department of Pathology & Laboratory Medicine, Anatomic Pathology, UCSF, San Francisco, CA, USA; 9University Cancer Center Leipzig (UCCL), Leipzig, Germany; 10Current address: Molecular Immunology, UCL Institute of Child Health, London, UK. Autologous human keratinocytes (HK) forming sheet and a strictly regulated spatiotemporal terminal differentiation grafts are approved as skin substitutes. Genetic engineer- program, which ultimately leads to the formation of the stratum ing of HK represents a promising technique to improve corneum, representing a water impermeable layer of avital corni- engraftment and survival of transplants.