Catalytic Receptors
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Supporting Information
Supporting Information Celhar et al. 10.1073/pnas.1507052112 SI Materials and Methods using a Nanodrop spectrophotometer (Thermo Fisher Scien- Proteinuria. Proteinuria was assessed using Albustix (Bayer). Al- tific). A TaqMan RNA-to-CT 1-Step Kit (Applied Biosystems) bumin levels in urine were assayed using an Albumin Mouse was used to perform the reverse transcription and quantitative ELISA Kit (Abcam) according to the manufacturer’s instructions; PCR reactions according to the manufacturer’s instructions samples were assayed at a dilution of 1:400. Samples were nor- using TaqMan gene expression assays (Applied Biosystems) to malized for creatinine using a Creatinine (urinary) Colorimetric either Tlr7 (Mm00446590) or the B2m housekeeping gene Assay Kit (Cayman Chemical) according to the manufacturer’s (Mm00437762). Real-time PCR was performed on the 7900H instructions; initial sample dilution of 1:10. fast real-time PCR system and analyzed using SDS 2.4 (Applied Biosystems). Relative mRNA expression was calculated using the Cell Sorting, RNA Isolation, and RT-PCR. Splenic B cells were comparative C method. + − + + t sorted as live CD45 Gr1 B220 CD19 , splenic T cells as live + − + + CD45 Gr1 CD3 CD5 and peritoneal macrophages as live Imaging. Kidney sections from OCT embedded tissue were fixed + − CD45 Gr1 CD11bhiF4/80hi. Sorted cells were centrifuged, re- with 4% paraformaldehyde before permeabilization with acetone suspended in TRIzol (Life Technologies) and stored at −80°. RNA and stained with Phalloidin (AF647) and anti-CD3d (unlabeled was extracted by TRIzol/chloroform and purified with the Qiagen Ab followed by secondary staining with donkey anti-goat Dylight RNeasy Mini purification kit according to the manufacturer’s 550). -

Molecular Mechanisms Involved Involved in the Interaction Effects of HCV and Ethanol on Liver Cirrhosis
Virginia Commonwealth University VCU Scholars Compass Theses and Dissertations Graduate School 2010 Molecular Mechanisms Involved Involved in the Interaction Effects of HCV and Ethanol on Liver Cirrhosis Ryan Fassnacht Virginia Commonwealth University Follow this and additional works at: https://scholarscompass.vcu.edu/etd Part of the Physiology Commons © The Author Downloaded from https://scholarscompass.vcu.edu/etd/2246 This Thesis is brought to you for free and open access by the Graduate School at VCU Scholars Compass. It has been accepted for inclusion in Theses and Dissertations by an authorized administrator of VCU Scholars Compass. For more information, please contact [email protected]. Ryan C. Fassnacht 2010 All Rights Reserved Molecular Mechanisms Involved in the Interaction Effects of HCV and Ethanol on Liver Cirrhosis A thesis submitted in partial fulfillment of the requirements for the degree of Master of Science at Virginia Commonwealth University. by Ryan Christopher Fassnacht, B.S. Hampden Sydney University, 2005 M.S. Virginia Commonwealth University, 2010 Director: Valeria Mas, Ph.D., Associate Professor of Surgery and Pathology Division of Transplant Department of Surgery Virginia Commonwealth University Richmond, Virginia July 9, 2010 Acknowledgement The Author wishes to thank his family and close friends for their support. He would also like to thank the members of the molecular transplant team for their help and advice. This project would not have been possible with out the help of Dr. Valeria Mas and her endearing -

Cytokine Nomenclature
RayBiotech, Inc. The protein array pioneer company Cytokine Nomenclature Cytokine Name Official Full Name Genbank Related Names Symbol 4-1BB TNFRSF Tumor necrosis factor NP_001552 CD137, ILA, 4-1BB ligand receptor 9 receptor superfamily .2. member 9 6Ckine CCL21 6-Cysteine Chemokine NM_002989 Small-inducible cytokine A21, Beta chemokine exodus-2, Secondary lymphoid-tissue chemokine, SLC, SCYA21 ACE ACE Angiotensin-converting NP_000780 CD143, DCP, DCP1 enzyme .1. NP_690043 .1. ACE-2 ACE2 Angiotensin-converting NP_068576 ACE-related carboxypeptidase, enzyme 2 .1 Angiotensin-converting enzyme homolog ACTH ACTH Adrenocorticotropic NP_000930 POMC, Pro-opiomelanocortin, hormone .1. Corticotropin-lipotropin, NPP, NP_001030 Melanotropin gamma, Gamma- 333.1 MSH, Potential peptide, Corticotropin, Melanotropin alpha, Alpha-MSH, Corticotropin-like intermediary peptide, CLIP, Lipotropin beta, Beta-LPH, Lipotropin gamma, Gamma-LPH, Melanotropin beta, Beta-MSH, Beta-endorphin, Met-enkephalin ACTHR ACTHR Adrenocorticotropic NP_000520 Melanocortin receptor 2, MC2-R hormone receptor .1 Activin A INHBA Activin A NM_002192 Activin beta-A chain, Erythroid differentiation protein, EDF, INHBA Activin B INHBB Activin B NM_002193 Inhibin beta B chain, Activin beta-B chain Activin C INHBC Activin C NM005538 Inhibin, beta C Activin RIA ACVR1 Activin receptor type-1 NM_001105 Activin receptor type I, ACTR-I, Serine/threonine-protein kinase receptor R1, SKR1, Activin receptor-like kinase 2, ALK-2, TGF-B superfamily receptor type I, TSR-I, ACVRLK2 Activin RIB ACVR1B -

Meconium Ileus Caused by Mutations in GUCY2C, Encoding the CFTR-Activating Guanylate Cyclase 2C
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Elsevier - Publisher Connector REPORT Meconium Ileus Caused by Mutations in GUCY2C, Encoding the CFTR-Activating Guanylate Cyclase 2C Hila Romi,1,6 Idan Cohen,1,6 Daniella Landau,2 Suliman Alkrinawi,2 Baruch Yerushalmi,2 Reli Hershkovitz,3 Nitza Newman-Heiman,2 Garry R. Cutting,4 Rivka Ofir,1 Sara Sivan,1 and Ohad S. Birk1,5,* Meconium ileus, intestinal obstruction in the newborn, is caused in most cases by CFTR mutations modulated by yet-unidentified modi- fier genes. We now show that in two unrelated consanguineous Bedouin kindreds, an autosomal-recessive phenotype of meconium ileus that is not associated with cystic fibrosis (CF) is caused by different homozygous mutations in GUCY2C, leading to a dramatic reduction or fully abrogating the enzymatic activity of the encoded guanlyl cyclase 2C. GUCY2C is a transmembrane receptor whose extracellular domain is activated by either the endogenous ligands, guanylin and related peptide uroguanylin, or by an external ligand, Escherichia coli (E. coli) heat-stable enterotoxin STa. GUCY2C is expressed in the human intestine, and the encoded protein activates the CFTR protein through local generation of cGMP. Thus, GUCY2C is a likely candidate modifier of the meconium ileus phenotype in CF. Because GUCY2C heterozygous and homozygous mutant mice are resistant to E. coli STa enterotoxin-induced diarrhea, it is plausible that GUCY2C mutations in the desert-dwelling Bedouin kindred are of selective advantage. Meconium ileus (MI), intestinal obstruction by inspissated homozygosity on chromosome 12p13 (spanning 9.5 Mb meconium in the distal ileum and cecum, develops in between markers D12S366 and D12S310) that was utero and presents shortly after birth as failure to pass common to all affected individuals. -

Supporting Online Material
1 2 3 4 5 6 7 Supplementary Information for 8 9 Fractalkine-induced microglial vasoregulation occurs within the retina and is altered early in diabetic 10 retinopathy 11 12 *Samuel A. Mills, *Andrew I. Jobling, *Michael A. Dixon, Bang V. Bui, Kirstan A. Vessey, Joanna A. Phipps, 13 Ursula Greferath, Gene Venables, Vickie H.Y. Wong, Connie H.Y. Wong, Zheng He, Flora Hui, James C. 14 Young, Josh Tonc, Elena Ivanova, Botir T. Sagdullaev, Erica L. Fletcher 15 * Joint first authors 16 17 Corresponding author: 18 Prof. Erica L. Fletcher. Department of Anatomy & Neuroscience. The University of Melbourne, Grattan St, 19 Parkville 3010, Victoria, Australia. 20 Email: [email protected] ; Tel: +61-3-8344-3218; Fax: +61-3-9347-5219 21 22 This PDF file includes: 23 24 Supplementary text 25 Figures S1 to S10 26 Tables S1 to S7 27 Legends for Movies S1 to S2 28 SI References 29 30 Other supplementary materials for this manuscript include the following: 31 32 Movies S1 to S2 33 34 35 36 1 1 Supplementary Information Text 2 Materials and Methods 3 Microglial process movement on retinal vessels 4 Dark agouti rats were anaesthetized, injected intraperitoneally with rhodamine B (Sigma-Aldrich) to label blood 5 vessels and retinal explants established as described in the main text. Retinal microglia were labelled with Iba-1 6 and imaging performed on an inverted confocal microscope (Leica SP5). Baseline images were taken for 10 7 minutes, followed by the addition of PBS (10 minutes) and then either fractalkine or fractalkine + candesartan 8 (10 minutes) using concentrations outlined in the main text. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Supp Table 1.Pdf
Upregulated genes in Hdac8 null cranial neural crest cells fold change Gene Symbol Gene Title 134.39 Stmn4 stathmin-like 4 46.05 Lhx1 LIM homeobox protein 1 31.45 Lect2 leukocyte cell-derived chemotaxin 2 31.09 Zfp108 zinc finger protein 108 27.74 0710007G10Rik RIKEN cDNA 0710007G10 gene 26.31 1700019O17Rik RIKEN cDNA 1700019O17 gene 25.72 Cyb561 Cytochrome b-561 25.35 Tsc22d1 TSC22 domain family, member 1 25.27 4921513I08Rik RIKEN cDNA 4921513I08 gene 24.58 Ofa oncofetal antigen 24.47 B230112I24Rik RIKEN cDNA B230112I24 gene 23.86 Uty ubiquitously transcribed tetratricopeptide repeat gene, Y chromosome 22.84 D8Ertd268e DNA segment, Chr 8, ERATO Doi 268, expressed 19.78 Dag1 Dystroglycan 1 19.74 Pkn1 protein kinase N1 18.64 Cts8 cathepsin 8 18.23 1500012D20Rik RIKEN cDNA 1500012D20 gene 18.09 Slc43a2 solute carrier family 43, member 2 17.17 Pcm1 Pericentriolar material 1 17.17 Prg2 proteoglycan 2, bone marrow 17.11 LOC671579 hypothetical protein LOC671579 17.11 Slco1a5 solute carrier organic anion transporter family, member 1a5 17.02 Fbxl7 F-box and leucine-rich repeat protein 7 17.02 Kcns2 K+ voltage-gated channel, subfamily S, 2 16.93 AW493845 Expressed sequence AW493845 16.12 1600014K23Rik RIKEN cDNA 1600014K23 gene 15.71 Cst8 cystatin 8 (cystatin-related epididymal spermatogenic) 15.68 4922502D21Rik RIKEN cDNA 4922502D21 gene 15.32 2810011L19Rik RIKEN cDNA 2810011L19 gene 15.08 Btbd9 BTB (POZ) domain containing 9 14.77 Hoxa11os homeo box A11, opposite strand transcript 14.74 Obp1a odorant binding protein Ia 14.72 ORF28 open reading -

Cellular and Molecular Signatures in the Disease Tissue of Early
Cellular and Molecular Signatures in the Disease Tissue of Early Rheumatoid Arthritis Stratify Clinical Response to csDMARD-Therapy and Predict Radiographic Progression Frances Humby1,* Myles Lewis1,* Nandhini Ramamoorthi2, Jason Hackney3, Michael Barnes1, Michele Bombardieri1, Francesca Setiadi2, Stephen Kelly1, Fabiola Bene1, Maria di Cicco1, Sudeh Riahi1, Vidalba Rocher-Ros1, Nora Ng1, Ilias Lazorou1, Rebecca E. Hands1, Desiree van der Heijde4, Robert Landewé5, Annette van der Helm-van Mil4, Alberto Cauli6, Iain B. McInnes7, Christopher D. Buckley8, Ernest Choy9, Peter Taylor10, Michael J. Townsend2 & Costantino Pitzalis1 1Centre for Experimental Medicine and Rheumatology, William Harvey Research Institute, Barts and The London School of Medicine and Dentistry, Queen Mary University of London, Charterhouse Square, London EC1M 6BQ, UK. Departments of 2Biomarker Discovery OMNI, 3Bioinformatics and Computational Biology, Genentech Research and Early Development, South San Francisco, California 94080 USA 4Department of Rheumatology, Leiden University Medical Center, The Netherlands 5Department of Clinical Immunology & Rheumatology, Amsterdam Rheumatology & Immunology Center, Amsterdam, The Netherlands 6Rheumatology Unit, Department of Medical Sciences, Policlinico of the University of Cagliari, Cagliari, Italy 7Institute of Infection, Immunity and Inflammation, University of Glasgow, Glasgow G12 8TA, UK 8Rheumatology Research Group, Institute of Inflammation and Ageing (IIA), University of Birmingham, Birmingham B15 2WB, UK 9Institute of -

Angiopoietin 4 (ANGPT4) (NM 015985) Human Untagged Clone Product Data
OriGene Technologies, Inc. 9620 Medical Center Drive, Ste 200 Rockville, MD 20850, US Phone: +1-888-267-4436 [email protected] EU: [email protected] CN: [email protected] Product datasheet for SC304373 Angiopoietin 4 (ANGPT4) (NM_015985) Human Untagged Clone Product data: Product Type: Expression Plasmids Product Name: Angiopoietin 4 (ANGPT4) (NM_015985) Human Untagged Clone Tag: Tag Free Symbol: ANGPT4 Synonyms: ANG3; ANG4 Vector: pCMV6-XL5 E. coli Selection: Ampicillin (100 ug/mL) Cell Selection: None This product is to be used for laboratory only. Not for diagnostic or therapeutic use. View online » ©2021 OriGene Technologies, Inc., 9620 Medical Center Drive, Ste 200, Rockville, MD 20850, US 1 / 3 Angiopoietin 4 (ANGPT4) (NM_015985) Human Untagged Clone – SC304373 Fully Sequenced ORF: >OriGene sequence for NM_015985 edited CAGGCAAGCCTGGCCACTGTTGGCTGCAGCAGGACATCCCAGGCACAGCCCCTAGGGCTC TGAGCAGACATCCCTCGCCATTGACACATCTTCAGATGCTCTCCCAGCTAGCCATGCTGC AGGGCAGCCTCCTCCTTGTGGTTGCCACCATGTCTGTGGCTCAACAGACAAGGCAGGAGG CGGATAGGGGCTGCGAGACACTTGTAGTCCAGCACGGCCACTGTAGCTACACCTTCTTGC TGCCCAAGTCTGAGCCCTGCCCTCCGGGGCCTGAGGTCTCCAGGGACTCCAACACCCTCC AGAGAGAATCACTGGCCAACCCACTGCACCTGGGGAAGTTGCCCACCCAGCAGGTGAAAC AGCTGGAGCAGGCACTGCAGAACAACACGCAGTGGCTGAAGAAGCTAGAGAGGGCCATCA AGACGATCTTGAGGTCGAAGCTGGAGCAGGTCCAGCAGCAAATGGCCCAGAATCAGACGG CCCCCATGCTAGAGCTGGGCACCAGCCTCCTGAACCAGACCACTGCCCAGATCCGCAAGC TGACCGACATGGAGGCTCAGCTCCTGAACCAGACATCAAGAATGGATGCCCAGATGCCAG AGACCTTTCTGTCCACCAACAAGCTGGAGAACCAGCTGCTGCTACAGAGGCAGAAGCTCC AGCAGCTTCAGGGCCAAAACAGCGCGCTCGAGAAGCGGTTGCAGGCCCTGGAGACCAAGC -
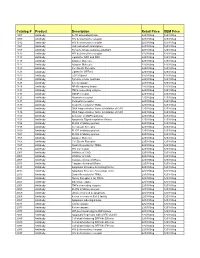
Comprehensive Product List
Catalog # Product Description Retail Price OEM Price 1007 Antibody IL-1R associated kinase 225/100ug 125/100ug 1009 Antibody HIV & chemokine receptor 225/100ug 125/100ug 1012 Antibody HIV & chemokine receptor 225/100ug 125/100ug 1021 Antibody JAK activated transcription 225/100ug 125/100ug 1107 Antibody Tyrosine kinase substrate p62DOK 225/100ug 125/100ug 1112 Antibody HIV & chemokine receptor 225/100ug 125/100ug 1113 Antibody Ligand for DR4 and DR5 225/100ug 125/100ug 1115 Antibody Adapter Molecule 225/100ug 125/100ug 1117 Antibody Adapter Molecule 225/100ug 125/100ug 1120 Antibody Cell Death Receptor 225/100ug 125/100ug 1121 Antibody Ligand for GFRa-2 225/100ug 125/100ug 1123 Antibody CCR3 ligand 225/100ug 125/100ug 1125 Antibody Tyrosine kinase substrate 225/100ug 125/100ug 1128 Antibody A new caspase 225/100ug 125/100ug 1129 Antibody NF-kB inducing kinase 225/100ug 125/100ug 1131 Antibody TNFa converting enzyme 225/100ug 125/100ug 1133 Antibody GDNF receptor 225/100ug 125/100ug 1135 Antibody Neurturin receptor 225/100ug 125/100ug 1137 Antibody Persephin receptor 225/100ug 125/100ug 1139 Antibody Death Receptor for TRAIL 225/100ug 125/100ug 1141 Antibody DNA fragmentation factor & Inhibitor of CAD 225/100ug 125/100ug 1148 Antibody DNA fragmentation factor & Inhibitor of CAD 225/100ug 125/100ug 1150 Antibody Activator of MAPK pathway 225/100ug 125/100ug 1151 Antibody Apoptosis Signal-regulation Kinase 225/100ug 125/100ug 1156 Antibody FLICE inhibitory protein 225/100ug 125/100ug 1158 Antibody Cell Death Receptor 225/100ug 125/100ug 1159 -

1714 Gene Comprehensive Cancer Panel Enriched for Clinically Actionable Genes with Additional Biologically Relevant Genes 400-500X Average Coverage on Tumor
xO GENE PANEL 1714 gene comprehensive cancer panel enriched for clinically actionable genes with additional biologically relevant genes 400-500x average coverage on tumor Genes A-C Genes D-F Genes G-I Genes J-L AATK ATAD2B BTG1 CDH7 CREM DACH1 EPHA1 FES G6PC3 HGF IL18RAP JADE1 LMO1 ABCA1 ATF1 BTG2 CDK1 CRHR1 DACH2 EPHA2 FEV G6PD HIF1A IL1R1 JAK1 LMO2 ABCB1 ATM BTG3 CDK10 CRK DAXX EPHA3 FGF1 GAB1 HIF1AN IL1R2 JAK2 LMO7 ABCB11 ATR BTK CDK11A CRKL DBH EPHA4 FGF10 GAB2 HIST1H1E IL1RAP JAK3 LMTK2 ABCB4 ATRX BTRC CDK11B CRLF2 DCC EPHA5 FGF11 GABPA HIST1H3B IL20RA JARID2 LMTK3 ABCC1 AURKA BUB1 CDK12 CRTC1 DCUN1D1 EPHA6 FGF12 GALNT12 HIST1H4E IL20RB JAZF1 LPHN2 ABCC2 AURKB BUB1B CDK13 CRTC2 DCUN1D2 EPHA7 FGF13 GATA1 HLA-A IL21R JMJD1C LPHN3 ABCG1 AURKC BUB3 CDK14 CRTC3 DDB2 EPHA8 FGF14 GATA2 HLA-B IL22RA1 JMJD4 LPP ABCG2 AXIN1 C11orf30 CDK15 CSF1 DDIT3 EPHB1 FGF16 GATA3 HLF IL22RA2 JMJD6 LRP1B ABI1 AXIN2 CACNA1C CDK16 CSF1R DDR1 EPHB2 FGF17 GATA5 HLTF IL23R JMJD7 LRP5 ABL1 AXL CACNA1S CDK17 CSF2RA DDR2 EPHB3 FGF18 GATA6 HMGA1 IL2RA JMJD8 LRP6 ABL2 B2M CACNB2 CDK18 CSF2RB DDX3X EPHB4 FGF19 GDNF HMGA2 IL2RB JUN LRRK2 ACE BABAM1 CADM2 CDK19 CSF3R DDX5 EPHB6 FGF2 GFI1 HMGCR IL2RG JUNB LSM1 ACSL6 BACH1 CALR CDK2 CSK DDX6 EPOR FGF20 GFI1B HNF1A IL3 JUND LTK ACTA2 BACH2 CAMTA1 CDK20 CSNK1D DEK ERBB2 FGF21 GFRA4 HNF1B IL3RA JUP LYL1 ACTC1 BAG4 CAPRIN2 CDK3 CSNK1E DHFR ERBB3 FGF22 GGCX HNRNPA3 IL4R KAT2A LYN ACVR1 BAI3 CARD10 CDK4 CTCF DHH ERBB4 FGF23 GHR HOXA10 IL5RA KAT2B LZTR1 ACVR1B BAP1 CARD11 CDK5 CTCFL DIAPH1 ERCC1 FGF3 GID4 HOXA11 IL6R KAT5 ACVR2A -

Diverse Functional Autoantibodies in Patients with COVID-19
medRxiv preprint doi: https://doi.org/10.1101/2020.12.10.20247205; this version posted December 11, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted medRxiv a license to display the preprint in perpetuity. It is made available under a CC-BY-NC-ND 4.0 International license . Diverse Functional Autoantibodies in Patients with COVID-19 Eric Y. Wang1,*, Tianyang Mao1,*, Jon Klein1,*, Yile Dai1,*, John D. Huck1, Feimei Liu1, Neil S. Zheng1, Ting Zhou1, Benjamin Israelow1, Patrick Wong1, Carolina Lucas1, Julio Silva1, Ji Eun Oh1, Eric Song1, Emily S. Perotti1, Suzanne Fischer1, Melissa Campbell5, John B. Fournier5, Anne L. Wyllie3, Chantal B. F. Vogels3, Isabel M. Ott3, Chaney C. Kalinich3, Mary E. Petrone3, Anne E. Watkins3, Yale IMPACT Team¶, Charles Dela Cruz4, Shelli F. Farhadian5, Wade L. Schulz6,7, Nathan D. Grubaugh3, Albert I. Ko3,5, Akiko Iwasaki1,3,8,#, Aaron M. Ring1,2,# 1 Department of Immunobiology, Yale School of Medicine, New Haven, CT, USA 2 Department of Pharmacology, Yale School of Medicine, New Haven, CT, USA 3 Department of Epidemiology of Microbial Diseases, Yale School of Public Health, New Haven, CT, USA 4 Department of Medicine, Section of Pulmonary and Critical Care Medicine, Yale School of Medicine, New Haven, CT, USA 5 Department of Internal Medicine (Infectious Diseases), Yale School of Medicine, New Haven, CT, USA 6 Department of Laboratory Medicine, Yale School of Medicine, New Haven, CT, USA 7 Center for Outcomes Research and Evaluation, Yale-New Haven Hospital, New Haven, CT, USA 8 Howard Hughes Medical Institute, Chevy Chase, MD, USA * These authors contributed equally to this work ¶ A list of authors and their affiliations appears at the end of the paper # Correspondence: [email protected] (A.M.R.); [email protected] (A.I.) 1 NOTE: This preprint reports new research that has not been certified by peer review and should not be used to guide clinical practice.