Supporting Information
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Molecular Mechanisms Involved Involved in the Interaction Effects of HCV and Ethanol on Liver Cirrhosis
Virginia Commonwealth University VCU Scholars Compass Theses and Dissertations Graduate School 2010 Molecular Mechanisms Involved Involved in the Interaction Effects of HCV and Ethanol on Liver Cirrhosis Ryan Fassnacht Virginia Commonwealth University Follow this and additional works at: https://scholarscompass.vcu.edu/etd Part of the Physiology Commons © The Author Downloaded from https://scholarscompass.vcu.edu/etd/2246 This Thesis is brought to you for free and open access by the Graduate School at VCU Scholars Compass. It has been accepted for inclusion in Theses and Dissertations by an authorized administrator of VCU Scholars Compass. For more information, please contact [email protected]. Ryan C. Fassnacht 2010 All Rights Reserved Molecular Mechanisms Involved in the Interaction Effects of HCV and Ethanol on Liver Cirrhosis A thesis submitted in partial fulfillment of the requirements for the degree of Master of Science at Virginia Commonwealth University. by Ryan Christopher Fassnacht, B.S. Hampden Sydney University, 2005 M.S. Virginia Commonwealth University, 2010 Director: Valeria Mas, Ph.D., Associate Professor of Surgery and Pathology Division of Transplant Department of Surgery Virginia Commonwealth University Richmond, Virginia July 9, 2010 Acknowledgement The Author wishes to thank his family and close friends for their support. He would also like to thank the members of the molecular transplant team for their help and advice. This project would not have been possible with out the help of Dr. Valeria Mas and her endearing -

A Functional C-Terminal TRAF3-Binding Site in MAVS Participates in Positive and Negative Regulation of the IFN Antiviral Response
Cell Research (2011) 21:895-910. © 2011 IBCB, SIBS, CAS All rights reserved 1001-0602/11 $ 32.00 npg ORIGINAL ARTICLE www.nature.com/cr A functional C-terminal TRAF3-binding site in MAVS participates in positive and negative regulation of the IFN antiviral response Suzanne Paz1, 2, Myriam Vilasco3, Steven J Werden1, Meztli Arguello1, Deshanthe Joseph-Pillai1, 2, Tiejun Zhao1, Thi Lien-Anh Nguyen1, Qiang Sun1, Eliane F Meurs3, Rongtuan Lin1, 4, John Hiscott1, 2, 4 1Terry Fox Molecular Oncology Group, Lady Davis Institute, Jewish General Hospital, Montreal, Quebec H3T1E2, Canada; 2Department of Microbiology and Immunology, McGill University Montreal, Quebec H3A 2B4, Canada; 3Department of Virology, Unit of Hepacivirus and Innate Immunity, Pasteur Institute, Paris 75724 France; 4Department of Medicine, McGill University Montreal, Quebec H3A 2B4, Canada Recognition of viral RNA structures by the cytosolic sensor retinoic acid-inducible gene-I (RIG-I) results in the activation of signaling cascades that culminate with the generation of the type I interferon (IFN) antiviral response. Onset of antiviral and inflammatory responses to viral pathogens necessitates the regulated spatiotemporal recruitment of signaling adapters, kinases and transcriptional proteins to the mitochondrial antiviral signaling protein (MAVS). We previously demonstrated that the serine/threonine kinase IKKε is recruited to the C-terminal region of MAVS following Sendai or vesicular stomatitis virus (VSV) infection, mediated by Lys63-linked polyubiquitination of MAVS at Lys500, resulting in inhibition of downstream IFN signaling (Paz et al, Mol Cell Biol, 2009). In this study, we demonstrate that C-terminus of MAVS harbors a novel TRAF3-binding site in the aa450- 468 region of MAVS. -

Mechanism of Action Through an IFN Type I-Independent Responses To
Downloaded from http://www.jimmunol.org/ by guest on September 25, 2021 is online at: average * The Journal of Immunology , 12 of which you can access for free at: 2012; 188:3088-3098; Prepublished online 20 from submission to initial decision 4 weeks from acceptance to publication February 2012; doi: 10.4049/jimmunol.1101764 http://www.jimmunol.org/content/188/7/3088 MF59 and Pam3CSK4 Boost Adaptive Responses to Influenza Subunit Vaccine through an IFN Type I-Independent Mechanism of Action Elena Caproni, Elaine Tritto, Mario Cortese, Alessandro Muzzi, Flaviana Mosca, Elisabetta Monaci, Barbara Baudner, Anja Seubert and Ennio De Gregorio J Immunol cites 33 articles Submit online. Every submission reviewed by practicing scientists ? is published twice each month by Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts http://jimmunol.org/subscription http://www.jimmunol.org/content/suppl/2012/02/21/jimmunol.110176 4.DC1 This article http://www.jimmunol.org/content/188/7/3088.full#ref-list-1 Information about subscribing to The JI No Triage! Fast Publication! Rapid Reviews! 30 days* Why • • • Material References Permissions Email Alerts Subscription Supplementary The Journal of Immunology The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2012 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 -

Supplementary Table S1. Control Capsules (CC) and Study Capsules (CR) Composition
Supplementary Table S1. Control Capsules (CC) and Study Capsules (CR) composition CC Capsules Composition mg Bioactive compound Sunflower oil 1.100 Gelatin 318,019 Glycerin 118,750 Clorophill E-140 1,923 CAPSULE TOTAL WEIGHT 1.538,692 CR Capsules Composition mg Bioactive compound Shark liver oil 20% 750 150 mg alkylglycerols alkylglycerols (Gustav Heess) Rosemary Antioxidant 11,25 mg Diterpene Phenols extract, 25 % Diterpene 45 Phenols, Type Nº 027.020 (Rosmarinus officinalis L.) Glyceril-monoestearate 30 Sunflower oil 263 Soy lecithin VEROLEC 56 12 Gelatin 318,019 Glycerin 118,750 Clorophill E-140 1,923 CAPSULE TOTAL WEIGHT 1.538,692 Supplementary Table S2. Table of the selected genes and pathways analyzed in the study Pathway Gen Gene name Inflammation, IL1B Interleukin 1, Beta Immunomodulation TNF (TNFA) Tumor Necrosis Factor MAPK1 Mitogen-Activated Protein Kinase 1 PTK2B Protein Tyrosine Kinase 2 Beta STAT3 Signal Transducer Activator Of Transcription 3 JAK1 Janus Kinase 1 JAK3 Janus Kinase 3 NFKB Nuclear Factor Of Kappa Light Polypeptide Gene Enhancer In B-Cells 1 NLRP3 NLR Family, Pyrin Domain Containing 3 CCL2 (MCP-1) Chemokine (C-C Motif) Ligand 2 CXCR1 Chemokine (C-X-C Motif) Receptor 1 CSF2 Colony Stimulating Factor 2 (Granulocyte-Macrophage) CCL5(RANTES) Chemokine (C-C Motif) Ligand 5 CCR5 Chemokine (C-C Motif) Receptor 5 (Gene/Pseudogene) PLCG1 Phospholipase C, Gamma 1 PRKCD Protein Kinase C, Delta ADIPOQ Adiponectin, C1Q And Collagen Domain Containing BMP2 Bone Morphogenetic Protein 2 LIF Leukemia Inhibitory Factor TGFB2 -

Supplemental Materials ZNF281 Enhances Cardiac Reprogramming
Supplemental Materials ZNF281 enhances cardiac reprogramming by modulating cardiac and inflammatory gene expression Huanyu Zhou, Maria Gabriela Morales, Hisayuki Hashimoto, Matthew E. Dickson, Kunhua Song, Wenduo Ye, Min S. Kim, Hanspeter Niederstrasser, Zhaoning Wang, Beibei Chen, Bruce A. Posner, Rhonda Bassel-Duby and Eric N. Olson Supplemental Table 1; related to Figure 1. Supplemental Table 2; related to Figure 1. Supplemental Table 3; related to the “quantitative mRNA measurement” in Materials and Methods section. Supplemental Table 4; related to the “ChIP-seq, gene ontology and pathway analysis” and “RNA-seq” and gene ontology analysis” in Materials and Methods section. Supplemental Figure S1; related to Figure 1. Supplemental Figure S2; related to Figure 2. Supplemental Figure S3; related to Figure 3. Supplemental Figure S4; related to Figure 4. Supplemental Figure S5; related to Figure 6. Supplemental Table S1. Genes included in human retroviral ORF cDNA library. Gene Gene Gene Gene Gene Gene Gene Gene Symbol Symbol Symbol Symbol Symbol Symbol Symbol Symbol AATF BMP8A CEBPE CTNNB1 ESR2 GDF3 HOXA5 IL17D ADIPOQ BRPF1 CEBPG CUX1 ESRRA GDF6 HOXA6 IL17F ADNP BRPF3 CERS1 CX3CL1 ETS1 GIN1 HOXA7 IL18 AEBP1 BUD31 CERS2 CXCL10 ETS2 GLIS3 HOXB1 IL19 AFF4 C17ORF77 CERS4 CXCL11 ETV3 GMEB1 HOXB13 IL1A AHR C1QTNF4 CFL2 CXCL12 ETV7 GPBP1 HOXB5 IL1B AIMP1 C21ORF66 CHIA CXCL13 FAM3B GPER HOXB6 IL1F3 ALS2CR8 CBFA2T2 CIR1 CXCL14 FAM3D GPI HOXB7 IL1F5 ALX1 CBFA2T3 CITED1 CXCL16 FASLG GREM1 HOXB9 IL1F6 ARGFX CBFB CITED2 CXCL3 FBLN1 GREM2 HOXC4 IL1F7 -

Human Ifnα4 / Ifna4 / Interferon Alpha-4 Protein (Fc Tag)
Human IFNα4 / IFNa4 / Interferon alpha-4 Protein (Fc Tag) Catalog Number: 10336-H01H General Information SDS-PAGE: Gene Name Synonym: IFN-alpha4a; INFA4; MGC142200 Protein Construction: A DNA sequence encoding the mature form of human IFN-α4a (NP_066546.1) (Cys 23-Asp 189) was expressed with the N-terminal fused Fc region of human IgG1. Source: Human Expression Host: HEK293 Cells QC Testing Purity: > 97 % as determined by SDS-PAGE Bio Activity: Protein Description Measured in antiviral assays using WISH human amnion cells infected Interferon, alpha 4 (IFNA4) belongs to the alpha/beta interferon family. Two with vesicular stomatitis virus (VSV). The EC50 for this effect is 0.2-1 variants of IFNA4 (IFNA4a and IFNA4b) are known, which differ from each ng/mL. other by changes in their coding regions at nucleotide positions 220 and 410 and can be distinguished by selective restriction enzyme analysis. Endotoxin: Interferons are produced by macrophages, IFN-alpha have antiviral activities. Interferon stimulates the production of two enzymes: a protein < 1.0 EU per μg of the protein as determined by the LAL method kinase and an oligoadenylate synthetase. IFN-alpha, the first cytokine to be produced by recombinant DNA technology, has emerged as an important Stability: regulator of growth and differentiation, affecting cellular communication and Samples are stable for up to twelve months from date of receipt at -70 ℃ signal transduction pathways as well as immunological control. Originally discovered as an antiviral substance, the efficacy of IFN-alpha in malignant, Predicted N terminal: Glu viral, immunological, angiogenic, inflammatory, and fibrotic diseases suggests a spectrum of interrelated pathophysiologies. -

Supplementary Material
Supplementary Material Table S1: Significant downregulated KEGGs pathways identified by DAVID following exposure to five cinnamon- based phenylpropanoids (p < 0.05). p-value Term: Genes (Benjamini) Cytokine-cytokine receptor interaction: FASLG, TNFSF14, CXCL11, IL11, FLT3LG, CCL3L1, CCL3L3, CXCR6, XCR1, 2.43 × 105 RTEL1, CSF2RA, TNFRSF17, TNFRSF14, CCNL2, VEGFB, AMH, TNFRSF10B, INHBE, IFNB1, CCR3, VEGFA, CCR2, IL12A, CCL1, CCL3, CXCL5, TNFRSF25, CCR1, CSF1, CX3CL1, CCL7, CCL24, TNFRSF1B, IL12RB1, CCL21, FIGF, EPO, IL4, IL18R1, FLT1, TGFBR1, EDA2R, HGF, TNFSF8, KDR, LEP, GH2, CCL13, EPOR, XCL1, IFNA16, XCL2 Neuroactive ligand-receptor interaction: OPRM1, THRA, GRIK1, DRD2, GRIK2, TACR2, TACR1, GABRB1, LPAR4, 9.68 × 105 GRIK5, FPR1, PRSS1, GNRHR, FPR2, EDNRA, AGTR2, LTB4R, PRSS2, CNR1, S1PR4, CALCRL, TAAR5, GABRE, PTGER1, GABRG3, C5AR1, PTGER3, PTGER4, GABRA6, GABRA5, GRM1, PLG, LEP, CRHR1, GH2, GRM3, SSTR2, Chlorogenic acid Chlorogenic CHRM3, GRIA1, MC2R, P2RX2, TBXA2R, GHSR, HTR2C, TSHR, LHB, GLP1R, OPRD1 Hematopoietic cell lineage: IL4, CR1, CD8B, CSF1, FCER2, GYPA, ITGA2, IL11, GP9, FLT3LG, CD38, CD19, DNTT, 9.29 × 104 GP1BB, CD22, EPOR, CSF2RA, CD14, THPO, EPO, HLA-DRA, ITGA2B Cytokine-cytokine receptor interaction: IL6ST, IL21R, IL19, TNFSF15, CXCR3, IL15, CXCL11, TGFB1, IL11, FLT3LG, CXCL10, CCR10, XCR1, RTEL1, CSF2RA, IL21, CCNL2, VEGFB, CCR8, AMH, TNFRSF10C, IFNB1, PDGFRA, EDA, CXCL5, TNFRSF25, CSF1, IFNW1, CNTFR, CX3CL1, CCL5, TNFRSF4, CCL4, CCL27, CCL24, CCL25, CCL23, IFNA6, IFNA5, FIGF, EPO, AMHR2, IL2RA, FLT4, TGFBR2, EDA2R, -

Diverse Functional Autoantibodies in Patients with COVID-19
medRxiv preprint doi: https://doi.org/10.1101/2020.12.10.20247205; this version posted December 11, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted medRxiv a license to display the preprint in perpetuity. It is made available under a CC-BY-NC-ND 4.0 International license . Diverse Functional Autoantibodies in Patients with COVID-19 Eric Y. Wang1,*, Tianyang Mao1,*, Jon Klein1,*, Yile Dai1,*, John D. Huck1, Feimei Liu1, Neil S. Zheng1, Ting Zhou1, Benjamin Israelow1, Patrick Wong1, Carolina Lucas1, Julio Silva1, Ji Eun Oh1, Eric Song1, Emily S. Perotti1, Suzanne Fischer1, Melissa Campbell5, John B. Fournier5, Anne L. Wyllie3, Chantal B. F. Vogels3, Isabel M. Ott3, Chaney C. Kalinich3, Mary E. Petrone3, Anne E. Watkins3, Yale IMPACT Team¶, Charles Dela Cruz4, Shelli F. Farhadian5, Wade L. Schulz6,7, Nathan D. Grubaugh3, Albert I. Ko3,5, Akiko Iwasaki1,3,8,#, Aaron M. Ring1,2,# 1 Department of Immunobiology, Yale School of Medicine, New Haven, CT, USA 2 Department of Pharmacology, Yale School of Medicine, New Haven, CT, USA 3 Department of Epidemiology of Microbial Diseases, Yale School of Public Health, New Haven, CT, USA 4 Department of Medicine, Section of Pulmonary and Critical Care Medicine, Yale School of Medicine, New Haven, CT, USA 5 Department of Internal Medicine (Infectious Diseases), Yale School of Medicine, New Haven, CT, USA 6 Department of Laboratory Medicine, Yale School of Medicine, New Haven, CT, USA 7 Center for Outcomes Research and Evaluation, Yale-New Haven Hospital, New Haven, CT, USA 8 Howard Hughes Medical Institute, Chevy Chase, MD, USA * These authors contributed equally to this work ¶ A list of authors and their affiliations appears at the end of the paper # Correspondence: [email protected] (A.M.R.); [email protected] (A.I.) 1 NOTE: This preprint reports new research that has not been certified by peer review and should not be used to guide clinical practice. -

Supplementary Material DNA Methylation in Inflammatory Pathways Modifies the Association Between BMI and Adult-Onset Non- Atopic
Supplementary Material DNA Methylation in Inflammatory Pathways Modifies the Association between BMI and Adult-Onset Non- Atopic Asthma Ayoung Jeong 1,2, Medea Imboden 1,2, Akram Ghantous 3, Alexei Novoloaca 3, Anne-Elie Carsin 4,5,6, Manolis Kogevinas 4,5,6, Christian Schindler 1,2, Gianfranco Lovison 7, Zdenko Herceg 3, Cyrille Cuenin 3, Roel Vermeulen 8, Deborah Jarvis 9, André F. S. Amaral 9, Florian Kronenberg 10, Paolo Vineis 11,12 and Nicole Probst-Hensch 1,2,* 1 Swiss Tropical and Public Health Institute, 4051 Basel, Switzerland; [email protected] (A.J.); [email protected] (M.I.); [email protected] (C.S.) 2 Department of Public Health, University of Basel, 4001 Basel, Switzerland 3 International Agency for Research on Cancer, 69372 Lyon, France; [email protected] (A.G.); [email protected] (A.N.); [email protected] (Z.H.); [email protected] (C.C.) 4 ISGlobal, Barcelona Institute for Global Health, 08003 Barcelona, Spain; [email protected] (A.-E.C.); [email protected] (M.K.) 5 Universitat Pompeu Fabra (UPF), 08002 Barcelona, Spain 6 CIBER Epidemiología y Salud Pública (CIBERESP), 08005 Barcelona, Spain 7 Department of Economics, Business and Statistics, University of Palermo, 90128 Palermo, Italy; [email protected] 8 Environmental Epidemiology Division, Utrecht University, Institute for Risk Assessment Sciences, 3584CM Utrecht, Netherlands; [email protected] 9 Population Health and Occupational Disease, National Heart and Lung Institute, Imperial College, SW3 6LR London, UK; [email protected] (D.J.); [email protected] (A.F.S.A.) 10 Division of Genetic Epidemiology, Medical University of Innsbruck, 6020 Innsbruck, Austria; [email protected] 11 MRC-PHE Centre for Environment and Health, School of Public Health, Imperial College London, W2 1PG London, UK; [email protected] 12 Italian Institute for Genomic Medicine (IIGM), 10126 Turin, Italy * Correspondence: [email protected]; Tel.: +41-61-284-8378 Int. -

Prevalent Homozygous Deletions of Type I Interferon and Defensin Genes in Human Cancers Associate with Immunotherapy Resistance
Author Manuscript Published OnlineFirst on April 4, 2018; DOI: 10.1158/1078-0432.CCR-17-3008 Author manuscripts have been peer reviewed and accepted for publication but have not yet been edited. Prevalent Homozygous Deletions of Type I Interferon and Defensin Genes in Human Cancers Associate with Immunotherapy Resistance Zhenqing Ye1; Haidong Dong2,3; Ying Li1; Tao Ma1,4; Haojie Huang4; Hon-Sing Leong4; Jeanette Eckel-Passow1; Jean-Pierre A. Kocher1; Han Liang5; LiguoWang1,4,* 1Division of Biomedical Statistics and Informatics, Department of Health Sciences, Mayo Clinic, Rochester, Minnesota 55905, United States of America 2Department of Immunology, College of Medicine, Mayo Clinic, Rochester, Minnesota 55905, United States of America 3Department of Urology, Mayo Clinic, Rochester, Minnesota 55905, United States of America 4Department of Biochemistry and Molecular Biology, Mayo Clinic, Rochester, Minnesota 55905, United States of America 5Department of Bioinformatics and Computational Biology, The University of Texas MD Anderson Cancer Center, Houston, Texas 77030, United States of America Running title: Pervasive Deletion of Interferon/Defensin in Human Cancers Keywords: Homozygous deletion, Type-I interferon, Defensin, immunotherapy resistance, cancer * Correspondence to: Liguo Wang, Ph.D. Associate Professor Division of Biomedical Statistics and Informatics, Mayo Clinic 200 1st St SW Rochester, MN 55905, USA Phone: +1-507-284-8728 Fax: +1-507-284-0745 Email: [email protected] The authors declare no potential conflicts of interest. 1 Downloaded from clincancerres.aacrjournals.org on September 29, 2021. © 2018 American Association for Cancer Research. Author Manuscript Published OnlineFirst on April 4, 2018; DOI: 10.1158/1078-0432.CCR-17-3008 Author manuscripts have been peer reviewed and accepted for publication but have not yet been edited. -
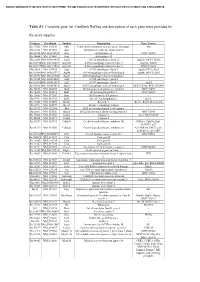
Table S1. Complete Gene List. Genbank Refseq and Description of Each Gene Were Provided By
Document downloaded from http://www.elsevier.es, day 24/09/2021. This copy is for personal use. Any transmission of this document by any media or format is strictly prohibited. Table S1. Complete gene list. GenBank RefSeq and description of each gene were provided by the array supplier. Unigene GeneBank Symbol Description Gene Name/s Rn.11422 NM_033230 Akt1 V-akt murine thymoma viral oncogene homolog 1 Akt Rn.2104 NM_019288 App Amyloid beta (A4) precursor protein - Rn.23323 NM_001034933 Arsa Arylsulfatase A MGC125207 Rn.94004 NM_033443 Arsb Arylsulfatase B - Rn.6224 NM_001038495 Atg12 ATG12 autophagy related 12 Apg12l, MGC125080 Rn.101734NM_001108809 Atg16l1 ATG16 autophagy related 16-like 1 Apg16l, Wdr30 Rn.104199NM_001191560 Atg16l2 ATG16 autophagy related 16-like 2 RGD1311400 Rn.3084 NM_134394 Atg3 ATG3 autophagy related 3 Apg3l, PIG-1, Pig1 Rn.163086NM_001025711 Atg4b ATG4 autophagy related 4 homolog B Apg4b, MGC112887 Rn.23378 NM_001107948 Atg4c ATG4 autophagy related 4 homolog C - Rn.98385 NM_001014250 Atg5 ATG5 autophagy related 5 - Rn.162765NM_001012097 Atg7 ATG7 autophagy related 7 Apg7l Rn.35248 NM_001014218 Atg9a ATG9 autophagy related 9 homolog A MGC105908, RGD1310450 Rn.36696 NM_022698 Bad BCL2-associated agonist of cell death MGC72439 Rn.14598 NM_053812 Bak1 BCL2-antagonist/killer 1 MGC108627 Rn.10668 NM_017059 Bax Bcl2-associated X protein - Rn.9996 NM_016993 Bcl2 B-cell CLL/lymphoma 2 Bcl-2 Rn.10323 NM_031535 Bcl2l1 Bcl2-like 1 Bcl-xl, Bcl2l, Bclx, bcl-X Rn.2776 NM_053739 Becn1 Beclin 1, autophagy related - Rn.31142 NM_022684 -

Single Cell Derived Clonal Analysis of Human Glioblastoma Links
SUPPLEMENTARY INFORMATION: Single cell derived clonal analysis of human glioblastoma links functional and genomic heterogeneity ! Mona Meyer*, Jüri Reimand*, Xiaoyang Lan, Renee Head, Xueming Zhu, Michelle Kushida, Jane Bayani, Jessica C. Pressey, Anath Lionel, Ian D. Clarke, Michael Cusimano, Jeremy Squire, Stephen Scherer, Mark Bernstein, Melanie A. Woodin, Gary D. Bader**, and Peter B. Dirks**! ! * These authors contributed equally to this work.! ** Correspondence: [email protected] or [email protected]! ! Supplementary information - Meyer, Reimand et al. Supplementary methods" 4" Patient samples and fluorescence activated cell sorting (FACS)! 4! Differentiation! 4! Immunocytochemistry and EdU Imaging! 4! Proliferation! 5! Western blotting ! 5! Temozolomide treatment! 5! NCI drug library screen! 6! Orthotopic injections! 6! Immunohistochemistry on tumor sections! 6! Promoter methylation of MGMT! 6! Fluorescence in situ Hybridization (FISH)! 7! SNP6 microarray analysis and genome segmentation! 7! Calling copy number alterations! 8! Mapping altered genome segments to genes! 8! Recurrently altered genes with clonal variability! 9! Global analyses of copy number alterations! 9! Phylogenetic analysis of copy number alterations! 10! Microarray analysis! 10! Gene expression differences of TMZ resistant and sensitive clones of GBM-482! 10! Reverse transcription-PCR analyses! 11! Tumor subtype analysis of TMZ-sensitive and resistant clones! 11! Pathway analysis of gene expression in the TMZ-sensitive clone of GBM-482! 11! Supplementary figures and tables" 13" "2 Supplementary information - Meyer, Reimand et al. Table S1: Individual clones from all patient tumors are tumorigenic. ! 14! Fig. S1: clonal tumorigenicity.! 15! Fig. S2: clonal heterogeneity of EGFR and PTEN expression.! 20! Fig. S3: clonal heterogeneity of proliferation.! 21! Fig.