Cutting Edge: Endogenous IFN-Β Regulates Survival and Development of Transitional B Cells
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Supporting Information
Supporting Information Celhar et al. 10.1073/pnas.1507052112 SI Materials and Methods using a Nanodrop spectrophotometer (Thermo Fisher Scien- Proteinuria. Proteinuria was assessed using Albustix (Bayer). Al- tific). A TaqMan RNA-to-CT 1-Step Kit (Applied Biosystems) bumin levels in urine were assayed using an Albumin Mouse was used to perform the reverse transcription and quantitative ELISA Kit (Abcam) according to the manufacturer’s instructions; PCR reactions according to the manufacturer’s instructions samples were assayed at a dilution of 1:400. Samples were nor- using TaqMan gene expression assays (Applied Biosystems) to malized for creatinine using a Creatinine (urinary) Colorimetric either Tlr7 (Mm00446590) or the B2m housekeeping gene Assay Kit (Cayman Chemical) according to the manufacturer’s (Mm00437762). Real-time PCR was performed on the 7900H instructions; initial sample dilution of 1:10. fast real-time PCR system and analyzed using SDS 2.4 (Applied Biosystems). Relative mRNA expression was calculated using the Cell Sorting, RNA Isolation, and RT-PCR. Splenic B cells were comparative C method. + − + + t sorted as live CD45 Gr1 B220 CD19 , splenic T cells as live + − + + CD45 Gr1 CD3 CD5 and peritoneal macrophages as live Imaging. Kidney sections from OCT embedded tissue were fixed + − CD45 Gr1 CD11bhiF4/80hi. Sorted cells were centrifuged, re- with 4% paraformaldehyde before permeabilization with acetone suspended in TRIzol (Life Technologies) and stored at −80°. RNA and stained with Phalloidin (AF647) and anti-CD3d (unlabeled was extracted by TRIzol/chloroform and purified with the Qiagen Ab followed by secondary staining with donkey anti-goat Dylight RNeasy Mini purification kit according to the manufacturer’s 550). -

Type I Interferons and the Development of Impaired Vascular Function and Repair in Human and Murine Lupus
Type I Interferons and the Development of Impaired Vascular Function and Repair in Human and Murine Lupus by Seth G Thacker A dissertation submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy (Immunology) in The University of Michigan 2011 Doctoral Committee: Associate Professor Mariana J. Kaplan, Chair Professor David A. Fox Professor Alisa E. Koch Professor Matthias Kretzler Professor Nicholas W. Lukacs Associate Professor Daniel T. Eitzman © Seth G Thacker 2011 Sharon, this work is dedicated to you. This achievement is as much yours as it is mine. Your support through all six years of this Ph.D. process has been incredible. You put up with my countless miscalculations on when I would finish experiments, and still managed to make me and our kids feel loved and special. Without you this would have no meaning. Sharon, you are the safe harbor in my life. ii Acknowledgments I have been exceptionally fortunate in my time here at the University of Michigan. I have been able to interact with so many supportive people over the years. I would like to express my thanks and admiration for my mentor. Mariana has taught me so much about writing, experimental design and being a successful scientist in general. I could never have made it here without her help. I would also like to thank Mike Denny. He had a hand in the beginning of all of my projects in one way or another, and was always quick and eager to help in whatever way he could. He really made my first year in the lab successful. -

Molecular Mechanisms Involved Involved in the Interaction Effects of HCV and Ethanol on Liver Cirrhosis
Virginia Commonwealth University VCU Scholars Compass Theses and Dissertations Graduate School 2010 Molecular Mechanisms Involved Involved in the Interaction Effects of HCV and Ethanol on Liver Cirrhosis Ryan Fassnacht Virginia Commonwealth University Follow this and additional works at: https://scholarscompass.vcu.edu/etd Part of the Physiology Commons © The Author Downloaded from https://scholarscompass.vcu.edu/etd/2246 This Thesis is brought to you for free and open access by the Graduate School at VCU Scholars Compass. It has been accepted for inclusion in Theses and Dissertations by an authorized administrator of VCU Scholars Compass. For more information, please contact [email protected]. Ryan C. Fassnacht 2010 All Rights Reserved Molecular Mechanisms Involved in the Interaction Effects of HCV and Ethanol on Liver Cirrhosis A thesis submitted in partial fulfillment of the requirements for the degree of Master of Science at Virginia Commonwealth University. by Ryan Christopher Fassnacht, B.S. Hampden Sydney University, 2005 M.S. Virginia Commonwealth University, 2010 Director: Valeria Mas, Ph.D., Associate Professor of Surgery and Pathology Division of Transplant Department of Surgery Virginia Commonwealth University Richmond, Virginia July 9, 2010 Acknowledgement The Author wishes to thank his family and close friends for their support. He would also like to thank the members of the molecular transplant team for their help and advice. This project would not have been possible with out the help of Dr. Valeria Mas and her endearing -

A Functional C-Terminal TRAF3-Binding Site in MAVS Participates in Positive and Negative Regulation of the IFN Antiviral Response
Cell Research (2011) 21:895-910. © 2011 IBCB, SIBS, CAS All rights reserved 1001-0602/11 $ 32.00 npg ORIGINAL ARTICLE www.nature.com/cr A functional C-terminal TRAF3-binding site in MAVS participates in positive and negative regulation of the IFN antiviral response Suzanne Paz1, 2, Myriam Vilasco3, Steven J Werden1, Meztli Arguello1, Deshanthe Joseph-Pillai1, 2, Tiejun Zhao1, Thi Lien-Anh Nguyen1, Qiang Sun1, Eliane F Meurs3, Rongtuan Lin1, 4, John Hiscott1, 2, 4 1Terry Fox Molecular Oncology Group, Lady Davis Institute, Jewish General Hospital, Montreal, Quebec H3T1E2, Canada; 2Department of Microbiology and Immunology, McGill University Montreal, Quebec H3A 2B4, Canada; 3Department of Virology, Unit of Hepacivirus and Innate Immunity, Pasteur Institute, Paris 75724 France; 4Department of Medicine, McGill University Montreal, Quebec H3A 2B4, Canada Recognition of viral RNA structures by the cytosolic sensor retinoic acid-inducible gene-I (RIG-I) results in the activation of signaling cascades that culminate with the generation of the type I interferon (IFN) antiviral response. Onset of antiviral and inflammatory responses to viral pathogens necessitates the regulated spatiotemporal recruitment of signaling adapters, kinases and transcriptional proteins to the mitochondrial antiviral signaling protein (MAVS). We previously demonstrated that the serine/threonine kinase IKKε is recruited to the C-terminal region of MAVS following Sendai or vesicular stomatitis virus (VSV) infection, mediated by Lys63-linked polyubiquitination of MAVS at Lys500, resulting in inhibition of downstream IFN signaling (Paz et al, Mol Cell Biol, 2009). In this study, we demonstrate that C-terminus of MAVS harbors a novel TRAF3-binding site in the aa450- 468 region of MAVS. -

Supplemental Materials ZNF281 Enhances Cardiac Reprogramming
Supplemental Materials ZNF281 enhances cardiac reprogramming by modulating cardiac and inflammatory gene expression Huanyu Zhou, Maria Gabriela Morales, Hisayuki Hashimoto, Matthew E. Dickson, Kunhua Song, Wenduo Ye, Min S. Kim, Hanspeter Niederstrasser, Zhaoning Wang, Beibei Chen, Bruce A. Posner, Rhonda Bassel-Duby and Eric N. Olson Supplemental Table 1; related to Figure 1. Supplemental Table 2; related to Figure 1. Supplemental Table 3; related to the “quantitative mRNA measurement” in Materials and Methods section. Supplemental Table 4; related to the “ChIP-seq, gene ontology and pathway analysis” and “RNA-seq” and gene ontology analysis” in Materials and Methods section. Supplemental Figure S1; related to Figure 1. Supplemental Figure S2; related to Figure 2. Supplemental Figure S3; related to Figure 3. Supplemental Figure S4; related to Figure 4. Supplemental Figure S5; related to Figure 6. Supplemental Table S1. Genes included in human retroviral ORF cDNA library. Gene Gene Gene Gene Gene Gene Gene Gene Symbol Symbol Symbol Symbol Symbol Symbol Symbol Symbol AATF BMP8A CEBPE CTNNB1 ESR2 GDF3 HOXA5 IL17D ADIPOQ BRPF1 CEBPG CUX1 ESRRA GDF6 HOXA6 IL17F ADNP BRPF3 CERS1 CX3CL1 ETS1 GIN1 HOXA7 IL18 AEBP1 BUD31 CERS2 CXCL10 ETS2 GLIS3 HOXB1 IL19 AFF4 C17ORF77 CERS4 CXCL11 ETV3 GMEB1 HOXB13 IL1A AHR C1QTNF4 CFL2 CXCL12 ETV7 GPBP1 HOXB5 IL1B AIMP1 C21ORF66 CHIA CXCL13 FAM3B GPER HOXB6 IL1F3 ALS2CR8 CBFA2T2 CIR1 CXCL14 FAM3D GPI HOXB7 IL1F5 ALX1 CBFA2T3 CITED1 CXCL16 FASLG GREM1 HOXB9 IL1F6 ARGFX CBFB CITED2 CXCL3 FBLN1 GREM2 HOXC4 IL1F7 -

Human Ifnα4 / Ifna4 / Interferon Alpha-4 Protein (Fc Tag)
Human IFNα4 / IFNa4 / Interferon alpha-4 Protein (Fc Tag) Catalog Number: 10336-H01H General Information SDS-PAGE: Gene Name Synonym: IFN-alpha4a; INFA4; MGC142200 Protein Construction: A DNA sequence encoding the mature form of human IFN-α4a (NP_066546.1) (Cys 23-Asp 189) was expressed with the N-terminal fused Fc region of human IgG1. Source: Human Expression Host: HEK293 Cells QC Testing Purity: > 97 % as determined by SDS-PAGE Bio Activity: Protein Description Measured in antiviral assays using WISH human amnion cells infected Interferon, alpha 4 (IFNA4) belongs to the alpha/beta interferon family. Two with vesicular stomatitis virus (VSV). The EC50 for this effect is 0.2-1 variants of IFNA4 (IFNA4a and IFNA4b) are known, which differ from each ng/mL. other by changes in their coding regions at nucleotide positions 220 and 410 and can be distinguished by selective restriction enzyme analysis. Endotoxin: Interferons are produced by macrophages, IFN-alpha have antiviral activities. Interferon stimulates the production of two enzymes: a protein < 1.0 EU per μg of the protein as determined by the LAL method kinase and an oligoadenylate synthetase. IFN-alpha, the first cytokine to be produced by recombinant DNA technology, has emerged as an important Stability: regulator of growth and differentiation, affecting cellular communication and Samples are stable for up to twelve months from date of receipt at -70 ℃ signal transduction pathways as well as immunological control. Originally discovered as an antiviral substance, the efficacy of IFN-alpha in malignant, Predicted N terminal: Glu viral, immunological, angiogenic, inflammatory, and fibrotic diseases suggests a spectrum of interrelated pathophysiologies. -

Cellular and Molecular Signatures in the Disease Tissue of Early
Cellular and Molecular Signatures in the Disease Tissue of Early Rheumatoid Arthritis Stratify Clinical Response to csDMARD-Therapy and Predict Radiographic Progression Frances Humby1,* Myles Lewis1,* Nandhini Ramamoorthi2, Jason Hackney3, Michael Barnes1, Michele Bombardieri1, Francesca Setiadi2, Stephen Kelly1, Fabiola Bene1, Maria di Cicco1, Sudeh Riahi1, Vidalba Rocher-Ros1, Nora Ng1, Ilias Lazorou1, Rebecca E. Hands1, Desiree van der Heijde4, Robert Landewé5, Annette van der Helm-van Mil4, Alberto Cauli6, Iain B. McInnes7, Christopher D. Buckley8, Ernest Choy9, Peter Taylor10, Michael J. Townsend2 & Costantino Pitzalis1 1Centre for Experimental Medicine and Rheumatology, William Harvey Research Institute, Barts and The London School of Medicine and Dentistry, Queen Mary University of London, Charterhouse Square, London EC1M 6BQ, UK. Departments of 2Biomarker Discovery OMNI, 3Bioinformatics and Computational Biology, Genentech Research and Early Development, South San Francisco, California 94080 USA 4Department of Rheumatology, Leiden University Medical Center, The Netherlands 5Department of Clinical Immunology & Rheumatology, Amsterdam Rheumatology & Immunology Center, Amsterdam, The Netherlands 6Rheumatology Unit, Department of Medical Sciences, Policlinico of the University of Cagliari, Cagliari, Italy 7Institute of Infection, Immunity and Inflammation, University of Glasgow, Glasgow G12 8TA, UK 8Rheumatology Research Group, Institute of Inflammation and Ageing (IIA), University of Birmingham, Birmingham B15 2WB, UK 9Institute of -

Supplementary Material
Supplementary Material Table S1: Significant downregulated KEGGs pathways identified by DAVID following exposure to five cinnamon- based phenylpropanoids (p < 0.05). p-value Term: Genes (Benjamini) Cytokine-cytokine receptor interaction: FASLG, TNFSF14, CXCL11, IL11, FLT3LG, CCL3L1, CCL3L3, CXCR6, XCR1, 2.43 × 105 RTEL1, CSF2RA, TNFRSF17, TNFRSF14, CCNL2, VEGFB, AMH, TNFRSF10B, INHBE, IFNB1, CCR3, VEGFA, CCR2, IL12A, CCL1, CCL3, CXCL5, TNFRSF25, CCR1, CSF1, CX3CL1, CCL7, CCL24, TNFRSF1B, IL12RB1, CCL21, FIGF, EPO, IL4, IL18R1, FLT1, TGFBR1, EDA2R, HGF, TNFSF8, KDR, LEP, GH2, CCL13, EPOR, XCL1, IFNA16, XCL2 Neuroactive ligand-receptor interaction: OPRM1, THRA, GRIK1, DRD2, GRIK2, TACR2, TACR1, GABRB1, LPAR4, 9.68 × 105 GRIK5, FPR1, PRSS1, GNRHR, FPR2, EDNRA, AGTR2, LTB4R, PRSS2, CNR1, S1PR4, CALCRL, TAAR5, GABRE, PTGER1, GABRG3, C5AR1, PTGER3, PTGER4, GABRA6, GABRA5, GRM1, PLG, LEP, CRHR1, GH2, GRM3, SSTR2, Chlorogenic acid Chlorogenic CHRM3, GRIA1, MC2R, P2RX2, TBXA2R, GHSR, HTR2C, TSHR, LHB, GLP1R, OPRD1 Hematopoietic cell lineage: IL4, CR1, CD8B, CSF1, FCER2, GYPA, ITGA2, IL11, GP9, FLT3LG, CD38, CD19, DNTT, 9.29 × 104 GP1BB, CD22, EPOR, CSF2RA, CD14, THPO, EPO, HLA-DRA, ITGA2B Cytokine-cytokine receptor interaction: IL6ST, IL21R, IL19, TNFSF15, CXCR3, IL15, CXCL11, TGFB1, IL11, FLT3LG, CXCL10, CCR10, XCR1, RTEL1, CSF2RA, IL21, CCNL2, VEGFB, CCR8, AMH, TNFRSF10C, IFNB1, PDGFRA, EDA, CXCL5, TNFRSF25, CSF1, IFNW1, CNTFR, CX3CL1, CCL5, TNFRSF4, CCL4, CCL27, CCL24, CCL25, CCL23, IFNA6, IFNA5, FIGF, EPO, AMHR2, IL2RA, FLT4, TGFBR2, EDA2R, -

WO 2018/067991 Al 12 April 2018 (12.04.2018) W !P O PCT
(12) INTERNATIONAL APPLICATION PUBLISHED UNDER THE PATENT COOPERATION TREATY (PCT) (19) World Intellectual Property Organization International Bureau (10) International Publication Number (43) International Publication Date WO 2018/067991 Al 12 April 2018 (12.04.2018) W !P O PCT (51) International Patent Classification: achusetts 021 15 (US). THE BROAD INSTITUTE, A61K 51/10 (2006.01) G01N 33/574 (2006.01) INC. [US/US]; 415 Main Street, Cambridge, Massachu C07K 14/705 (2006.01) A61K 47/68 (2017.01) setts 02142 (US). MASSACHUSETTS INSTITUTE OF G01N 33/53 (2006.01) TECHNOLOGY [US/US]; 77 Massachusetts Avenue, Cambridge, Massachusetts 02139 (US). (21) International Application Number: PCT/US2017/055625 (72) Inventors; and (71) Applicants: KUCHROO, Vijay K. [IN/US]; 30 Fairhaven (22) International Filing Date: Road, Newton, Massachusetts 02149 (US). ANDERSON, 06 October 2017 (06.10.2017) Ana Carrizosa [US/US]; 110 Cypress Street, Brookline, (25) Filing Language: English Massachusetts 02445 (US). MADI, Asaf [US/US]; c/o The Brigham and Women's Hospital, Inc., 75 Francis (26) Publication Language: English Street, Boston, Massachusetts 021 15 (US). CHIHARA, (30) Priority Data: Norio [US/US]; c/o The Brigham and Women's Hospital, 62/405,835 07 October 2016 (07.10.2016) US Inc., 75 Francis Street, Boston, Massachusetts 021 15 (US). REGEV, Aviv [US/US]; 15a Ellsworth Ave, Cambridge, (71) Applicants: THE BRIGHAM AND WOMEN'S HOSPI¬ Massachusetts 02139 (US). SINGER, Meromit [US/US]; TAL, INC. [US/US]; 75 Francis Street, Boston, Mass c/o The Broad Institute, Inc., 415 Main Street, Cambridge, (54) Title: MODULATION OF NOVEL IMMUNE CHECKPOINT TARGETS CD4 FIG. -

Supplementary Material DNA Methylation in Inflammatory Pathways Modifies the Association Between BMI and Adult-Onset Non- Atopic
Supplementary Material DNA Methylation in Inflammatory Pathways Modifies the Association between BMI and Adult-Onset Non- Atopic Asthma Ayoung Jeong 1,2, Medea Imboden 1,2, Akram Ghantous 3, Alexei Novoloaca 3, Anne-Elie Carsin 4,5,6, Manolis Kogevinas 4,5,6, Christian Schindler 1,2, Gianfranco Lovison 7, Zdenko Herceg 3, Cyrille Cuenin 3, Roel Vermeulen 8, Deborah Jarvis 9, André F. S. Amaral 9, Florian Kronenberg 10, Paolo Vineis 11,12 and Nicole Probst-Hensch 1,2,* 1 Swiss Tropical and Public Health Institute, 4051 Basel, Switzerland; [email protected] (A.J.); [email protected] (M.I.); [email protected] (C.S.) 2 Department of Public Health, University of Basel, 4001 Basel, Switzerland 3 International Agency for Research on Cancer, 69372 Lyon, France; [email protected] (A.G.); [email protected] (A.N.); [email protected] (Z.H.); [email protected] (C.C.) 4 ISGlobal, Barcelona Institute for Global Health, 08003 Barcelona, Spain; [email protected] (A.-E.C.); [email protected] (M.K.) 5 Universitat Pompeu Fabra (UPF), 08002 Barcelona, Spain 6 CIBER Epidemiología y Salud Pública (CIBERESP), 08005 Barcelona, Spain 7 Department of Economics, Business and Statistics, University of Palermo, 90128 Palermo, Italy; [email protected] 8 Environmental Epidemiology Division, Utrecht University, Institute for Risk Assessment Sciences, 3584CM Utrecht, Netherlands; [email protected] 9 Population Health and Occupational Disease, National Heart and Lung Institute, Imperial College, SW3 6LR London, UK; [email protected] (D.J.); [email protected] (A.F.S.A.) 10 Division of Genetic Epidemiology, Medical University of Innsbruck, 6020 Innsbruck, Austria; [email protected] 11 MRC-PHE Centre for Environment and Health, School of Public Health, Imperial College London, W2 1PG London, UK; [email protected] 12 Italian Institute for Genomic Medicine (IIGM), 10126 Turin, Italy * Correspondence: [email protected]; Tel.: +41-61-284-8378 Int. -

Prevalent Homozygous Deletions of Type I Interferon and Defensin Genes in Human Cancers Associate with Immunotherapy Resistance
Author Manuscript Published OnlineFirst on April 4, 2018; DOI: 10.1158/1078-0432.CCR-17-3008 Author manuscripts have been peer reviewed and accepted for publication but have not yet been edited. Prevalent Homozygous Deletions of Type I Interferon and Defensin Genes in Human Cancers Associate with Immunotherapy Resistance Zhenqing Ye1; Haidong Dong2,3; Ying Li1; Tao Ma1,4; Haojie Huang4; Hon-Sing Leong4; Jeanette Eckel-Passow1; Jean-Pierre A. Kocher1; Han Liang5; LiguoWang1,4,* 1Division of Biomedical Statistics and Informatics, Department of Health Sciences, Mayo Clinic, Rochester, Minnesota 55905, United States of America 2Department of Immunology, College of Medicine, Mayo Clinic, Rochester, Minnesota 55905, United States of America 3Department of Urology, Mayo Clinic, Rochester, Minnesota 55905, United States of America 4Department of Biochemistry and Molecular Biology, Mayo Clinic, Rochester, Minnesota 55905, United States of America 5Department of Bioinformatics and Computational Biology, The University of Texas MD Anderson Cancer Center, Houston, Texas 77030, United States of America Running title: Pervasive Deletion of Interferon/Defensin in Human Cancers Keywords: Homozygous deletion, Type-I interferon, Defensin, immunotherapy resistance, cancer * Correspondence to: Liguo Wang, Ph.D. Associate Professor Division of Biomedical Statistics and Informatics, Mayo Clinic 200 1st St SW Rochester, MN 55905, USA Phone: +1-507-284-8728 Fax: +1-507-284-0745 Email: [email protected] The authors declare no potential conflicts of interest. 1 Downloaded from clincancerres.aacrjournals.org on September 29, 2021. © 2018 American Association for Cancer Research. Author Manuscript Published OnlineFirst on April 4, 2018; DOI: 10.1158/1078-0432.CCR-17-3008 Author manuscripts have been peer reviewed and accepted for publication but have not yet been edited. -
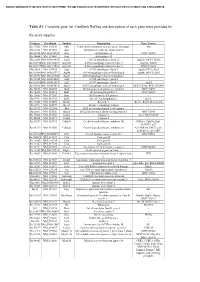
Table S1. Complete Gene List. Genbank Refseq and Description of Each Gene Were Provided By
Document downloaded from http://www.elsevier.es, day 24/09/2021. This copy is for personal use. Any transmission of this document by any media or format is strictly prohibited. Table S1. Complete gene list. GenBank RefSeq and description of each gene were provided by the array supplier. Unigene GeneBank Symbol Description Gene Name/s Rn.11422 NM_033230 Akt1 V-akt murine thymoma viral oncogene homolog 1 Akt Rn.2104 NM_019288 App Amyloid beta (A4) precursor protein - Rn.23323 NM_001034933 Arsa Arylsulfatase A MGC125207 Rn.94004 NM_033443 Arsb Arylsulfatase B - Rn.6224 NM_001038495 Atg12 ATG12 autophagy related 12 Apg12l, MGC125080 Rn.101734NM_001108809 Atg16l1 ATG16 autophagy related 16-like 1 Apg16l, Wdr30 Rn.104199NM_001191560 Atg16l2 ATG16 autophagy related 16-like 2 RGD1311400 Rn.3084 NM_134394 Atg3 ATG3 autophagy related 3 Apg3l, PIG-1, Pig1 Rn.163086NM_001025711 Atg4b ATG4 autophagy related 4 homolog B Apg4b, MGC112887 Rn.23378 NM_001107948 Atg4c ATG4 autophagy related 4 homolog C - Rn.98385 NM_001014250 Atg5 ATG5 autophagy related 5 - Rn.162765NM_001012097 Atg7 ATG7 autophagy related 7 Apg7l Rn.35248 NM_001014218 Atg9a ATG9 autophagy related 9 homolog A MGC105908, RGD1310450 Rn.36696 NM_022698 Bad BCL2-associated agonist of cell death MGC72439 Rn.14598 NM_053812 Bak1 BCL2-antagonist/killer 1 MGC108627 Rn.10668 NM_017059 Bax Bcl2-associated X protein - Rn.9996 NM_016993 Bcl2 B-cell CLL/lymphoma 2 Bcl-2 Rn.10323 NM_031535 Bcl2l1 Bcl2-like 1 Bcl-xl, Bcl2l, Bclx, bcl-X Rn.2776 NM_053739 Becn1 Beclin 1, autophagy related - Rn.31142 NM_022684