Data-Driven and Knowledge-Driven Computational Models of Angiogenesis in Application to Peripheral Arterial Disease
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Increased Expression of Epidermal Growth Factor Receptor and Betacellulin During the Early Stage of Gastric Ulcer Healing
505-510 11/6/08 12:55 Page 505 MOLECULAR MEDICINE REPORTS 1: 505-510, 2008 505 Increased expression of epidermal growth factor receptor and betacellulin during the early stage of gastric ulcer healing GEUN HAE CHOI, HO SUNG PARK, KYUNG RYOUL KIM, HA NA CHOI, KYU YUN JANG, MYOUNG JA CHUNG, MYOUNG JAE KANG, DONG GEUN LEE and WOO SUNG MOON Department of Pathology, Institute for Medical Sciences, Chonbuk National University Medical School and the Center for Healthcare Technology Development, Jeonju, Korea Received January 2, 2008; Accepted February 22, 2008 Abstract. Epidermal growth factor receptor (EGFR) is from tissue necrosis triggered by mucosal ischemia, free important for the proliferation and differentiation of gastric radical formation and the cessation of nutrient delivery, which mucosal cells. Betacellulin (BTC) is a novel ligand for EGFR are caused by vascular and microvascular injury such as Since their role is unclear in the ulcer healing process, we thrombi, constriction or other occlusions (2). Tissue necrosis investigated their expression. Gastric ulcers in 30 Sprague- and the release of leukotriene B attract leukocytes and Dawley rats were induced by acetic acid. RT-PCR and macrophages, which release pro-inflammatory cytokines Western blotting were performed to detect EGFR and BTC. (e.g. TNFα, IL-1α, and IL-1ß). These in turn activate local Immunohistochemical studies were performed to detect fibroblasts, endothelial and epithelial cells. Histologically, an EGFR, BTC and proliferating cell nuclear antigen (PCNA). ulcer has two characteristic structures: a distinct ulcer margin The expression of EGFR and the BTC gene was significantly formed by the adjacent non-necrotic mucosa, and granulation increased at 12 h, 24 h and 3 days after ulcer induction tissue composed of fibroblasts, macrophages and proliferating (P<0.05). -

Recombinant Human Betacellulin Promotes the Neogenesis of -Cells
Recombinant Human Betacellulin Promotes the Neogenesis of -Cells and Ameliorates Glucose Intolerance in Mice With Diabetes Induced by Selective Alloxan Perfusion Koji Yamamoto, Jun-ichiro Miyagawa, Masako Waguri, Reiko Sasada, Koichi Igarashi, Ming Li, Takao Nammo, Makoto Moriwaki, Akihisa Imagawa, Kazuya Yamagata, Hiromu Nakajima, Mitsuyoshi Namba, Yoshihiro Tochino, Toshiaki Hanafusa, and Yuji Matsuzawa Betacellulin (BTC), a member of the epidermal growth factor family, is expressed predominantly in the human pancreas and induces the differentiation of a pancreatic ancreatic -cells are thought to be terminally dif- acinar cell line (AR42J) into insulin-secreting cells, ferentiated cells with little ability to regenerate. suggesting that BTC has a physiologically important However, proliferation of preexisting -cells and role in the endocrine pancreas. In this study, we exam- differentiation of -cells from precursor cells, ined the in vivo effect of recombinant human BTC P (rhBTC) on glucose intolerance and pancreatic mor- mainly residing in the pancreatic duct lining, have been phology using a new mouse model with glucose intoler- demonstrated in some animal models (1–5). Recently, we ance induced by selective alloxan perfusion. RhBTC developed a new mouse model of diabetes induced by selec- (1 µg/g body wt) or saline was injected subcutaneously tive perfusion of alloxan (100 µg/g body wt) during the every day from the day after alloxan treatment. The clamping of the superior mesenteric artery (1). In this model, intraperitoneal glucose tolerance test revealed no dif- glucose intolerance spontaneously resolves after one year ference between rhBTC-treated and rhBTC-untreated because of the proliferation of surviving -cells in the non- glucose-intolerant mice at 2–4 weeks. -

Identification of the Binding Partners for Hspb2 and Cryab Reveals
Brigham Young University BYU ScholarsArchive Theses and Dissertations 2013-12-12 Identification of the Binding arP tners for HspB2 and CryAB Reveals Myofibril and Mitochondrial Protein Interactions and Non- Redundant Roles for Small Heat Shock Proteins Kelsey Murphey Langston Brigham Young University - Provo Follow this and additional works at: https://scholarsarchive.byu.edu/etd Part of the Microbiology Commons BYU ScholarsArchive Citation Langston, Kelsey Murphey, "Identification of the Binding Partners for HspB2 and CryAB Reveals Myofibril and Mitochondrial Protein Interactions and Non-Redundant Roles for Small Heat Shock Proteins" (2013). Theses and Dissertations. 3822. https://scholarsarchive.byu.edu/etd/3822 This Thesis is brought to you for free and open access by BYU ScholarsArchive. It has been accepted for inclusion in Theses and Dissertations by an authorized administrator of BYU ScholarsArchive. For more information, please contact [email protected], [email protected]. Identification of the Binding Partners for HspB2 and CryAB Reveals Myofibril and Mitochondrial Protein Interactions and Non-Redundant Roles for Small Heat Shock Proteins Kelsey Langston A thesis submitted to the faculty of Brigham Young University in partial fulfillment of the requirements for the degree of Master of Science Julianne H. Grose, Chair William R. McCleary Brian Poole Department of Microbiology and Molecular Biology Brigham Young University December 2013 Copyright © 2013 Kelsey Langston All Rights Reserved ABSTRACT Identification of the Binding Partners for HspB2 and CryAB Reveals Myofibril and Mitochondrial Protein Interactors and Non-Redundant Roles for Small Heat Shock Proteins Kelsey Langston Department of Microbiology and Molecular Biology, BYU Master of Science Small Heat Shock Proteins (sHSP) are molecular chaperones that play protective roles in cell survival and have been shown to possess chaperone activity. -

Aggf1 Attenuates Neuroinflammation and BBB Disruption Via PI3K/Akt/NF-Κb Pathway After Subarachnoid Hemorrhage in Rats
Zhu et al. Journal of Neuroinflammation (2018) 15:178 https://doi.org/10.1186/s12974-018-1211-8 RESEARCH Open Access Aggf1 attenuates neuroinflammation and BBB disruption via PI3K/Akt/NF-κB pathway after subarachnoid hemorrhage in rats Qiquan Zhu1,2, Budbazar Enkhjargal2, Lei Huang2,4, Tongyu Zhang2, Chengmei Sun2, Zhiyi Xie2, Pei Wu2, Jun Mo2, Jiping Tang2, Zongyi Xie1* and John H. Zhang2,3,4* Abstract Background: Neuroinflammation and blood-brain barrier (BBB) disruption are two critical mechanisms of subarachnoid hemorrhage (SAH)-induced brain injury, which are closely related to patient prognosis. Recently, angiogenic factor with G-patch and FHA domain 1 (Aggf1) was shown to inhibit inflammatory effect and preserve vascular integrity in non-nervous system diseases. This study aimed to determine whether Aggf1 could attenuate neuroinflammation and preserve BBB integrity after experimental SAH, as well as the underlying mechanisms of its protective roles. Methods: Two hundred forty-nine male Sprague-Dawley rats were subjected to the endovascular perforation model of SAH. Recombinant human Aggf1 (rh-Aggf1) was administered intravenously via tail vein injection at 1 h after SAH induction. To investigate the underlying neuroprotection mechanism, Aggf1 small interfering RNA (Aggf1 siRNA) and PI3K-specific inhibitor LY294002 were administered through intracerebroventricular (i.c.v.) before SAH induction. SAH grade, neurological score, brain water content, BBB permeability, Western blot, and immunohistochemistry were performed. Results: Expression of endogenous Aggf1 was markedly increased after SAH. Aggf1 was primarily expressed in endothelial cells and astrocytes, as well as microglia after SAH. Administration of rh-Aggf1 significantly reduced brain water content and BBB permeability, decreased the numbers of infiltrating neutrophils, and activated microglia in the ipsilateral cerebral cortex following SAH. -

Beta-Arrestin-Mediated Signaling in the Heart
SPECIAL ARTICLE Circ J 2008; 72: 1725–1729 Beta-Arrestin-Mediated Signaling in the Heart Priyesh A. Patel, BS; Douglas G. Tilley, PhD*; Howard A. Rockman, MD*,** Beta-arrestin is a multifunctional adapter protein well known for its role in G-protein-coupled receptor (GPCR) desensitization. Exciting new evidence indicates thatβ-arrestin is also a signaling molecule capable of initiating its own G-protein-independent signaling at GPCRs. One of the best-studiedβ-arrestin signaling pathways is the one involvingβ-arrestin-dependent activation of a mitogen-activated protein kinase cascade, the extracellular regulated kinase (ERK). ERK signaling, which is classically activated by agonist stimulation of the epidermal growth factor receptor (EGFR), can be activated by a number of GPCRs in aβ-arrestin-dependent manner. Recent work in animal models of heart failure suggests thatβ-arrestin-dependent activation of EGFR/ERK signaling by theβ-1-adrenergic receptor, and possibly the angiotensin II Type 1A receptor, are cardioprotective. Hence, a new model of signaling at cardiac GPCRs has emerged and implicates classical G-protein-mediated signaling with promoting harmful remodeling in heart failure, while concurrently linkingβ-arrestin-dependent, G-protein-inde- pendent signaling with cardioprotective effects. Based on this paradigm, a new class of drugs could be identified, termed “biased ligands”, which simultaneously block harmful G-protein signaling, while also promoting cardio- protectiveβ-arrestin-dependent signaling, leading to a potential breakthrough -

BMC Evolutionary Biology Biomed Central
BMC Evolutionary Biology BioMed Central Research article Open Access On the origins of arrestin and rhodopsin Carlos E Alvarez1,2,3 Address: 1Center for Molecular and Human Genetics, The Research Institute at Nationwide Children's Hospital, Columbus, OH, 43205, USA, 2Department of Pediatrics, The Ohio State University College of Medicine, Columbus, OH, 43210, USA and 3Novartis Institutes of BioMedical Research, CH-4002 Basel, Switzerland Email: Carlos E Alvarez - [email protected] Published: 29 July 2008 Received: 11 January 2008 Accepted: 29 July 2008 BMC Evolutionary Biology 2008, 8:222 doi:10.1186/1471-2148-8-222 This article is available from: http://www.biomedcentral.com/1471-2148/8/222 © 2008 Alvarez; licensee BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Abstract Background: G protein coupled receptors (GPCRs) are the most numerous proteins in mammalian genomes, and the most common targets of clinical drugs. However, their evolution remains enigmatic. GPCRs are intimately associated with trimeric G proteins, G protein receptor kinases, and arrestins. We conducted phylogenetic studies to reconstruct the history of arrestins. Those findings, in turn, led us to investigate the origin of the photosensory GPCR rhodopsin. Results: We found that the arrestin clan is comprised of the Spo0M protein family in archaea and bacteria, and the arrestin and Vps26 families in eukaryotes. The previously known animal arrestins are members of the visual/beta subfamily, which branched from the founding "alpha" arrestins relatively recently. -

The Role and Mechanisms of Action of Micrornas in Cancer Drug Resistance Wengong Si1,2,3, Jiaying Shen4, Huilin Zheng1,5 and Weimin Fan1,6*
Si et al. Clinical Epigenetics (2019) 11:25 https://doi.org/10.1186/s13148-018-0587-8 REVIEW Open Access The role and mechanisms of action of microRNAs in cancer drug resistance Wengong Si1,2,3, Jiaying Shen4, Huilin Zheng1,5 and Weimin Fan1,6* Abstract MicroRNAs (miRNAs) are small non-coding RNAs with a length of about 19–25 nt, which can regulate various target genes and are thus involved in the regulation of a variety of biological and pathological processes, including the formation and development of cancer. Drug resistance in cancer chemotherapy is one of the main obstacles to curing this malignant disease. Statistical data indicate that over 90% of the mortality of patients with cancer is related to drug resistance. Drug resistance of cancer chemotherapy can be caused by many mechanisms, such as decreased antitumor drug uptake, modified drug targets, altered cell cycle checkpoints, or increased DNA damage repair, among others. In recent years, many studies have shown that miRNAs are involved in the drug resistance of tumor cells by targeting drug-resistance-related genes or influencing genes related to cell proliferation, cell cycle, and apoptosis. A single miRNA often targets a number of genes, and its regulatory effect is tissue-specific. In this review, we emphasize the miRNAs that are involved in the regulation of drug resistance among different cancers and probe the mechanisms of the deregulated expression of miRNAs. The molecular targets of miRNAs and their underlying signaling pathways are also explored comprehensively. A holistic understanding of the functions of miRNAs in drug resistance will help us develop better strategies to regulate them efficiently and will finally pave the way toward better translation of miRNAs into clinics, developing them into a promising approach in cancer therapy. -

Supporting Online Material
1 2 3 4 5 6 7 Supplementary Information for 8 9 Fractalkine-induced microglial vasoregulation occurs within the retina and is altered early in diabetic 10 retinopathy 11 12 *Samuel A. Mills, *Andrew I. Jobling, *Michael A. Dixon, Bang V. Bui, Kirstan A. Vessey, Joanna A. Phipps, 13 Ursula Greferath, Gene Venables, Vickie H.Y. Wong, Connie H.Y. Wong, Zheng He, Flora Hui, James C. 14 Young, Josh Tonc, Elena Ivanova, Botir T. Sagdullaev, Erica L. Fletcher 15 * Joint first authors 16 17 Corresponding author: 18 Prof. Erica L. Fletcher. Department of Anatomy & Neuroscience. The University of Melbourne, Grattan St, 19 Parkville 3010, Victoria, Australia. 20 Email: [email protected] ; Tel: +61-3-8344-3218; Fax: +61-3-9347-5219 21 22 This PDF file includes: 23 24 Supplementary text 25 Figures S1 to S10 26 Tables S1 to S7 27 Legends for Movies S1 to S2 28 SI References 29 30 Other supplementary materials for this manuscript include the following: 31 32 Movies S1 to S2 33 34 35 36 1 1 Supplementary Information Text 2 Materials and Methods 3 Microglial process movement on retinal vessels 4 Dark agouti rats were anaesthetized, injected intraperitoneally with rhodamine B (Sigma-Aldrich) to label blood 5 vessels and retinal explants established as described in the main text. Retinal microglia were labelled with Iba-1 6 and imaging performed on an inverted confocal microscope (Leica SP5). Baseline images were taken for 10 7 minutes, followed by the addition of PBS (10 minutes) and then either fractalkine or fractalkine + candesartan 8 (10 minutes) using concentrations outlined in the main text. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Views of the NIH
CLINICAL EPIDEMIOLOGY www.jasn.org Genetic Variants Associated with Circulating Fibroblast Growth Factor 23 Cassianne Robinson-Cohen ,1 Traci M. Bartz,2 Dongbing Lai,3 T. Alp Ikizler,1 Munro Peacock,4 Erik A. Imel,4 Erin D. Michos,5 Tatiana M. Foroud,3 Kristina Akesson,6,7 Kent D. Taylor,8 Linnea Malmgren,6,7 Kunihiro Matsushita,5,9,10 Maria Nethander,11 Joel Eriksson,12 Claes Ohlsson,12 Daniel Mellström,12 Myles Wolf,13 Osten Ljunggren,14 Fiona McGuigan,6,7 Jerome I. Rotter,8 Magnus Karlsson,6,7 Michael J. Econs,3,4 Joachim H. Ix,15,16 Pamela L. Lutsey,17 Bruce M. Psaty,18,19 Ian H. de Boer ,20 and Bryan R. Kestenbaum 20 Due to the number of contributing authors, the affiliations are listed at the end of this article. ABSTRACT Background Fibroblast growth factor 23 (FGF23), a bone-derived hormone that regulates phosphorus and vitamin D metabolism, contributes to the pathogenesis of mineral and bone disorders in CKD and is an emerging cardiovascular risk factor. Central elements of FGF23 regulation remain incompletely under- stood; genetic variation may help explain interindividual differences. Methods We performed a meta-analysis of genome-wide association studies of circulating FGF23 con- centrations among 16,624 participants of European ancestry from seven cohort studies, excluding par- ticipants with eGFR,30 ml/min per 1.73 m2 to focus on FGF23 under normal conditions. We evaluated the association of single-nucleotide polymorphisms (SNPs) with natural log–transformed FGF23 concentra- tion, adjusted for age, sex, study site, and principal components of ancestry. -

Supplemental Table 1. Complete Gene Lists and GO Terms from Figure 3C
Supplemental Table 1. Complete gene lists and GO terms from Figure 3C. Path 1 Genes: RP11-34P13.15, RP4-758J18.10, VWA1, CHD5, AZIN2, FOXO6, RP11-403I13.8, ARHGAP30, RGS4, LRRN2, RASSF5, SERTAD4, GJC2, RHOU, REEP1, FOXI3, SH3RF3, COL4A4, ZDHHC23, FGFR3, PPP2R2C, CTD-2031P19.4, RNF182, GRM4, PRR15, DGKI, CHMP4C, CALB1, SPAG1, KLF4, ENG, RET, GDF10, ADAMTS14, SPOCK2, MBL1P, ADAM8, LRP4-AS1, CARNS1, DGAT2, CRYAB, AP000783.1, OPCML, PLEKHG6, GDF3, EMP1, RASSF9, FAM101A, STON2, GREM1, ACTC1, CORO2B, FURIN, WFIKKN1, BAIAP3, TMC5, HS3ST4, ZFHX3, NLRP1, RASD1, CACNG4, EMILIN2, L3MBTL4, KLHL14, HMSD, RP11-849I19.1, SALL3, GADD45B, KANK3, CTC- 526N19.1, ZNF888, MMP9, BMP7, PIK3IP1, MCHR1, SYTL5, CAMK2N1, PINK1, ID3, PTPRU, MANEAL, MCOLN3, LRRC8C, NTNG1, KCNC4, RP11, 430C7.5, C1orf95, ID2-AS1, ID2, GDF7, KCNG3, RGPD8, PSD4, CCDC74B, BMPR2, KAT2B, LINC00693, ZNF654, FILIP1L, SH3TC1, CPEB2, NPFFR2, TRPC3, RP11-752L20.3, FAM198B, TLL1, CDH9, PDZD2, CHSY3, GALNT10, FOXQ1, ATXN1, ID4, COL11A2, CNR1, GTF2IP4, FZD1, PAX5, RP11-35N6.1, UNC5B, NKX1-2, FAM196A, EBF3, PRRG4, LRP4, SYT7, PLBD1, GRASP, ALX1, HIP1R, LPAR6, SLITRK6, C16orf89, RP11-491F9.1, MMP2, B3GNT9, NXPH3, TNRC6C-AS1, LDLRAD4, NOL4, SMAD7, HCN2, PDE4A, KANK2, SAMD1, EXOC3L2, IL11, EMILIN3, KCNB1, DOK5, EEF1A2, A4GALT, ADGRG2, ELF4, ABCD1 Term Count % PValue Genes regulation of pathway-restricted GDF3, SMAD7, GDF7, BMPR2, GDF10, GREM1, BMP7, LDLRAD4, SMAD protein phosphorylation 9 6.34 1.31E-08 ENG pathway-restricted SMAD protein GDF3, SMAD7, GDF7, BMPR2, GDF10, GREM1, BMP7, LDLRAD4, phosphorylation -
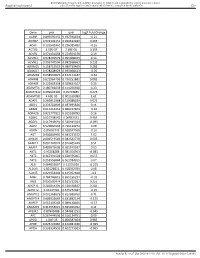
Gene Pval Qval Log2 Fold Change AAMP 0.895690332 0.952598834
BMJ Publishing Group Limited (BMJ) disclaims all liability and responsibility arising from any reliance Supplemental material placed on this supplemental material which has been supplied by the author(s) Gut Gene pval qval Log2 Fold Change AAMP 0.895690332 0.952598834 -0.21 ABI3BP 0.002302151 0.020612283 0.465 ACHE 0.103542461 0.296385483 -0.16 ACTG2 2.99E-07 7.68E-05 3.195 ACVR1 0.071431098 0.224504378 0.19 ACVR1C 0.978209579 0.995008423 0.14 ACVRL1 0.006747504 0.042938663 0.235 ADAM15 0.158715519 0.380719469 0.285 ADAM17 0.978208929 0.995008423 -0.05 ADAM28 0.038932876 0.152174187 -0.62 ADAM8 0.622964796 0.790251882 0.085 ADAM9 0.122003358 0.329623107 0.25 ADAMTS1 0.180766659 0.414256926 0.23 ADAMTS12 0.009902195 0.05703885 0.425 ADAMTS8 4.60E-05 0.001169089 1.61 ADAP1 0.269811968 0.519388039 0.075 ADD1 0.233702809 0.487695826 0.11 ADM2 0.012213453 0.066227879 -0.36 ADRA2B 0.822777921 0.915518785 0.16 AEBP1 0.010738542 0.06035531 0.465 AGGF1 0.117946691 0.320915024 -0.095 AGR2 0.529860903 0.736120272 0.08 AGRN 0.85693743 0.928047568 -0.16 AGT 0.006849995 0.043233572 1.02 AHNAK 0.006519543 0.042542779 0.605 AKAP12 0.001747074 0.016405449 0.51 AKAP2 0.409929603 0.665919397 0.05 AKT1 0.95208288 0.985354963 -0.085 AKT2 0.367391504 0.620376005 0.055 AKT3 0.253556844 0.501934205 0.07 ALB 0.064833867 0.21195036 -0.315 ALDOA 0.83128831 0.918352939 0.08 ALOX5 0.029954404 0.125352668 -0.3 AMH 0.784746815 0.895196237 -0.03 ANG 0.050500474 0.181732067 0.255 ANGPT1 0.281853305 0.538528647 0.285 ANGPT2 0.43147281 0.675272487 -0.15 ANGPTL2 0.001368876 0.013688762 0.71 ANGPTL4 0.686032669 0.831882134 -0.175 ANPEP 0.019103243 0.089148466 -0.57 ANXA2P2 0.412553021 0.665966092 0.11 AP1M2 0.87843088 0.944681253 -0.045 APC 0.267444505 0.516134751 0.09 APOD 1.04E-05 0.000587404 0.985 APOE 0.023722987 0.104981036 -0.395 APOH 0.336334555 0.602273505 -0.065 Sundar R, et al.