Differing from Diseased Stage in Melanoma, the Olfactory Receptor Gene OR56A4 Is Expressed
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Identification of Candidate Biomarkers and Pathways Associated with Type 1 Diabetes Mellitus Using Bioinformatics Analysis
bioRxiv preprint doi: https://doi.org/10.1101/2021.06.08.447531; this version posted June 9, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Identification of candidate biomarkers and pathways associated with type 1 diabetes mellitus using bioinformatics analysis Basavaraj Vastrad1, Chanabasayya Vastrad*2 1. Department of Biochemistry, Basaveshwar College of Pharmacy, Gadag, Karnataka 582103, India. 2. Biostatistics and Bioinformatics, Chanabasava Nilaya, Bharthinagar, Dharwad 580001, Karnataka, India. * Chanabasayya Vastrad [email protected] Ph: +919480073398 Chanabasava Nilaya, Bharthinagar, Dharwad 580001 , Karanataka, India bioRxiv preprint doi: https://doi.org/10.1101/2021.06.08.447531; this version posted June 9, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Abstract Type 1 diabetes mellitus (T1DM) is a metabolic disorder for which the underlying molecular mechanisms remain largely unclear. This investigation aimed to elucidate essential candidate genes and pathways in T1DM by integrated bioinformatics analysis. In this study, differentially expressed genes (DEGs) were analyzed using DESeq2 of R package from GSE162689 of the Gene Expression Omnibus (GEO). Gene ontology (GO) enrichment analysis, REACTOME pathway enrichment analysis, and construction and analysis of protein-protein interaction (PPI) network, modules, miRNA-hub gene regulatory network and TF-hub gene regulatory network, and validation of hub genes were then performed. A total of 952 DEGs (477 up regulated and 475 down regulated genes) were identified in T1DM. GO and REACTOME enrichment result results showed that DEGs mainly enriched in multicellular organism development, detection of stimulus, diseases of signal transduction by growth factor receptors and second messengers, and olfactory signaling pathway. -

WO 2019/068007 Al Figure 2
(12) INTERNATIONAL APPLICATION PUBLISHED UNDER THE PATENT COOPERATION TREATY (PCT) (19) World Intellectual Property Organization I International Bureau (10) International Publication Number (43) International Publication Date WO 2019/068007 Al 04 April 2019 (04.04.2019) W 1P O PCT (51) International Patent Classification: (72) Inventors; and C12N 15/10 (2006.01) C07K 16/28 (2006.01) (71) Applicants: GROSS, Gideon [EVIL]; IE-1-5 Address C12N 5/10 (2006.0 1) C12Q 1/6809 (20 18.0 1) M.P. Korazim, 1292200 Moshav Almagor (IL). GIBSON, C07K 14/705 (2006.01) A61P 35/00 (2006.01) Will [US/US]; c/o ImmPACT-Bio Ltd., 2 Ilian Ramon St., C07K 14/725 (2006.01) P.O. Box 4044, 7403635 Ness Ziona (TL). DAHARY, Dvir [EilL]; c/o ImmPACT-Bio Ltd., 2 Ilian Ramon St., P.O. (21) International Application Number: Box 4044, 7403635 Ness Ziona (IL). BEIMAN, Merav PCT/US2018/053583 [EilL]; c/o ImmPACT-Bio Ltd., 2 Ilian Ramon St., P.O. (22) International Filing Date: Box 4044, 7403635 Ness Ziona (E.). 28 September 2018 (28.09.2018) (74) Agent: MACDOUGALL, Christina, A. et al; Morgan, (25) Filing Language: English Lewis & Bockius LLP, One Market, Spear Tower, SanFran- cisco, CA 94105 (US). (26) Publication Language: English (81) Designated States (unless otherwise indicated, for every (30) Priority Data: kind of national protection available): AE, AG, AL, AM, 62/564,454 28 September 2017 (28.09.2017) US AO, AT, AU, AZ, BA, BB, BG, BH, BN, BR, BW, BY, BZ, 62/649,429 28 March 2018 (28.03.2018) US CA, CH, CL, CN, CO, CR, CU, CZ, DE, DJ, DK, DM, DO, (71) Applicant: IMMP ACT-BIO LTD. -
Quantitative-PCR Validation of 154 Genomic Segments Called As Cnvs in Five Replicat
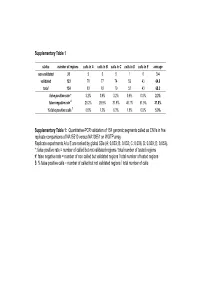
Supplementary Table 1 status number of regions calls in A calls in B calls in C calls in D calls in E average non validated 31 5 6 5 1 0 3.4 validated 123 78 77 74 52 43 64.8 total 154 83 83 79 53 43 68.2 false positive rate * 3.2% 3.9% 3.2% 0.6% 0.0% 2.2% false negative rate # 29.2% 29.9% 31.8% 46.1% 51.9% 37.8% % false positive calls $ 6.0% 7.2% 6.3% 1.9% 0.0% 5.0% Supplementary Table 1: Quantitative-PCR validation of 154 genomic segments called as CNVs in five replicate comparisons of NA15510 versus NA10851 on WGTP array Replicate experiments A to E are ranked by global SDe (A: 0.033; B: 0.033; C: 0.036; D: 0.039; E: 0.053). *: false positive rate = number of called but not validated regions / total number of tested regions #: false negative rate = number of non called but validated regions / total number of tested regions $: % false positive calls = number of called but not validated regions / total number of calls False positive estimates for 500K EA CNV calls Total Rep1 Rep2 Rep3 Avg (unique) Validated 33 28 32 31 38 Not validated 2 2 2 2 5 Total 35 30 34 33 43 % False positive 5.71% 6.67% 5.88% 6.09% - % False negative 13.16% 26.32% 15.79% 18.42% - Supplementary Table 2A : Quantitative PCR validation of 43 unique CNV regions called as CNVs in three replicate comparisons of NA15510 versus NA10851 using the 500K EA array. -

Table S4. RAE Analysis of Dedifferentiated Liposarcoma
Table S4. RAE analysis of dedifferentiated liposarcoma Model Chromosome Region start Region end Size q value freqX0* # genes genes Amp 1 57809872 60413476 2603605 0.00026 34.6 10 DAB1,RPS26P15,OMA1,TACSTD2,MYSM1,JUN,FGGY,HOOK1,CYP2J2,C1orf87 Amp 1 158619146 158696968 77823 0.053 25 1 VANGL2 Amp 1 158883523 158922841 39319 0.081 23.1 2 SLAMF1,CD48 Amp 1 162042586 162118557 75972 0.072 25 0 [Nearest:NUF2] Amp 1 162272460 162767627 495168 0.017 26.9 0 [Nearest:PBX1] Amp 1 165486554 165532374 45821 0.057 25 1 POU2F1 Amp 1 167138282 167483267 344986 0.024 26.9 2 ATP1B1,NME7 Amp 1 167612872 167708844 95973 0.041 25 3 BLZF1,C1orf114,SLC19A2 Amp 1 167728199 167808161 79963 0.076 21.2 1 F5 Amp 1 168436370 169233893 797524 0.018 26.9 3 GORAB,PRRX1,C1orf129 Amp 1 169462231 170768440 1306210 1.3E-06 38.5 10 FMO1,FMO4,TOP1P1,BAT2D1,MYOC,VAMP4,METTL13,DNM3,C1orf105,PIGC Amp 1 171026247 171291427 265181 0.015 26.9 1 TNFSF18 Del 1 201860394 202299299 438906 0.0047 25 6 ATP2B4,SNORA77,LAX1,ZC3H11A,SNRPE,C1orf157 Del 1 210909187 212021116 1111930 0.017 19.2 8 BATF3,NSL1,TATDN3,C1orf227,FLVCR1,VASH2,ANGEL2,RPS6KC1 Del 1 215937857 216049214 111358 0.079 23.1 1 SPATA17 Del 1 218237257 218367476 130220 0.0063 26.9 3 EPRS,BPNT1,IARS2 Del 1 222100886 222727238 626353 5.2E-05 32.7 5 FBXO28,DEGS1,NVL,CNIH4,WDR26 Del 1 223166548 224519805 1353258 0.0063 26.9 15 DNAH14,LBR,ENAH,SRP9,EPHX1,TMEM63A,LEFTY1,PYCR2,LEFTY2,C1orf55,H3F3A,LOC440926 ,ACBD3,MIXL1,LIN9 Del 1 225283136 225374166 91031 0.054 23.1 1 CDC42BPA Del 1 227278990 229012661 1733672 0.091 21.2 13 RAB4A,SPHAR,C1orf96,ACTA1,NUP133,ABCB10,TAF5L,URB2,GALNT2,PGBD5,COG2,AGT,CAP -

The Hypothalamus As a Hub for SARS-Cov-2 Brain Infection and Pathogenesis
bioRxiv preprint doi: https://doi.org/10.1101/2020.06.08.139329; this version posted June 19, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. The hypothalamus as a hub for SARS-CoV-2 brain infection and pathogenesis Sreekala Nampoothiri1,2#, Florent Sauve1,2#, Gaëtan Ternier1,2ƒ, Daniela Fernandois1,2 ƒ, Caio Coelho1,2, Monica ImBernon1,2, Eleonora Deligia1,2, Romain PerBet1, Vincent Florent1,2,3, Marc Baroncini1,2, Florence Pasquier1,4, François Trottein5, Claude-Alain Maurage1,2, Virginie Mattot1,2‡, Paolo GiacoBini1,2‡, S. Rasika1,2‡*, Vincent Prevot1,2‡* 1 Univ. Lille, Inserm, CHU Lille, Lille Neuroscience & Cognition, DistAlz, UMR-S 1172, Lille, France 2 LaBoratorY of Development and PlasticitY of the Neuroendocrine Brain, FHU 1000 daYs for health, EGID, School of Medicine, Lille, France 3 Nutrition, Arras General Hospital, Arras, France 4 Centre mémoire ressources et recherche, CHU Lille, LiCEND, Lille, France 5 Univ. Lille, CNRS, INSERM, CHU Lille, Institut Pasteur de Lille, U1019 - UMR 8204 - CIIL - Center for Infection and ImmunitY of Lille (CIIL), Lille, France. # and ƒ These authors contriButed equallY to this work. ‡ These authors directed this work *Correspondence to: [email protected] and [email protected] Short title: Covid-19: the hypothalamic hypothesis 1 bioRxiv preprint doi: https://doi.org/10.1101/2020.06.08.139329; this version posted June 19, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. -

DNA Copy Number Aberrations Associated with Aneuploidy and Chromosomal Instability in Breast Cancers
875-883.qxd 25/8/2010 10:49 Ì ™ÂÏ›‰·875 ONCOLOGY REPORTS 24: 875-883, 2010 875 DNA copy number aberrations associated with aneuploidy and chromosomal instability in breast cancers SHIGETO KAWAUCHI1, TOMOKO FURUYA1, KENZO IKEMOTO1, MOTONAO NAKAO1, SHIGERU YAMAMOTO2, MASAAKI OKA2 and KOHSUKE SASAKI1 Departments of 1Pathology and 2Surgery, Yamaguchi University Graduate School of Medicine, Ube, Yamaguchi 755-8505, Japan Received May 8, 2010; Accepted June 21, 2010 DOI: 10.3892/or_00000933 Abstract. Biological characteristics of a tumor are primarily somal material (1,2). Aneuploidy, an alteration in the number affected by its genomic alterations. It is thus important to of chromosomes or nuclear DNA content, is a common trait ascertain whether there are genomic changes linked with of tumor cells, and it drives tumor progression (3). With DNA ploidy and/or chromosomal instability (CIN). In the tumor progression, aneuploidy successively confers a more present study, using fresh-frozen samples of 46 invasive aggressive character to tumor cells (4). Thus, aneuploidy breast cancers, laser scanning cytometry, array-based com- has been used as a prognostic marker of various kinds of parative genomic hybridization, and chromosome fluore- cancers including breast cancer (5-7). scence in situ hybridization were performed to assess DNA Genomic instability is also recognized as inherent chara- ploidy, DNA copy number aberrations (DCNAs), and CIN cteristics of cancer cells. The underlying mechanisms of status. Both ploidy and CIN status were examined in 36 genomic instability are different among tumors. Genomic tumors, resulting in 23 aneuploid/CIN+ tumors, 1 aneuploid/ instability is roughly divided into microsatellite instability CIN-, 2 diploid/CIN+, and 10 diploid/CIN- tumors. -

Amino Acid Sequences Directed Against Cxcr4 And
(19) TZZ ¥¥_T (11) EP 2 285 833 B1 (12) EUROPEAN PATENT SPECIFICATION (45) Date of publication and mention (51) Int Cl.: of the grant of the patent: C07K 16/28 (2006.01) A61K 39/395 (2006.01) 17.12.2014 Bulletin 2014/51 A61P 31/18 (2006.01) A61P 35/00 (2006.01) (21) Application number: 09745851.7 (86) International application number: PCT/EP2009/056026 (22) Date of filing: 18.05.2009 (87) International publication number: WO 2009/138519 (19.11.2009 Gazette 2009/47) (54) AMINO ACID SEQUENCES DIRECTED AGAINST CXCR4 AND OTHER GPCRs AND COMPOUNDS COMPRISING THE SAME GEGEN CXCR4 UND ANDERE GPCR GERICHTETE AMINOSÄURESEQUENZEN SOWIE VERBINDUNGEN DAMIT SÉQUENCES D’ACIDES AMINÉS DIRIGÉES CONTRE CXCR4 ET AUTRES GPCR ET COMPOSÉS RENFERMANT CES DERNIÈRES (84) Designated Contracting States: (74) Representative: Hoffmann Eitle AT BE BG CH CY CZ DE DK EE ES FI FR GB GR Patent- und Rechtsanwälte PartmbB HR HU IE IS IT LI LT LU LV MC MK MT NL NO PL Arabellastraße 30 PT RO SE SI SK TR 81925 München (DE) (30) Priority: 16.05.2008 US 53847 P (56) References cited: 02.10.2008 US 102142 P EP-A- 1 316 801 WO-A-99/50461 WO-A-03/050531 WO-A-03/066830 (43) Date of publication of application: WO-A-2006/089141 WO-A-2007/051063 23.02.2011 Bulletin 2011/08 • VADAY GAYLE G ET AL: "CXCR4 and CXCL12 (73) Proprietor: Ablynx N.V. (SDF-1) in prostate cancer: inhibitory effects of 9052 Ghent-Zwijnaarde (BE) human single chain Fv antibodies" CLINICAL CANCER RESEARCH, THE AMERICAN (72) Inventors: ASSOCIATION FOR CANCER RESEARCH, US, • BLANCHETOT, Christophe vol.10, no. -

Us 2018 / 0305689 A1
US 20180305689A1 ( 19 ) United States (12 ) Patent Application Publication ( 10) Pub . No. : US 2018 /0305689 A1 Sætrom et al. ( 43 ) Pub . Date: Oct. 25 , 2018 ( 54 ) SARNA COMPOSITIONS AND METHODS OF plication No . 62 /150 , 895 , filed on Apr. 22 , 2015 , USE provisional application No . 62/ 150 ,904 , filed on Apr. 22 , 2015 , provisional application No. 62 / 150 , 908 , (71 ) Applicant: MINA THERAPEUTICS LIMITED , filed on Apr. 22 , 2015 , provisional application No. LONDON (GB ) 62 / 150 , 900 , filed on Apr. 22 , 2015 . (72 ) Inventors : Pål Sætrom , Trondheim (NO ) ; Endre Publication Classification Bakken Stovner , Trondheim (NO ) (51 ) Int . CI. C12N 15 / 113 (2006 .01 ) (21 ) Appl. No. : 15 /568 , 046 (52 ) U . S . CI. (22 ) PCT Filed : Apr. 21 , 2016 CPC .. .. .. C12N 15 / 113 ( 2013 .01 ) ; C12N 2310 / 34 ( 2013. 01 ) ; C12N 2310 /14 (2013 . 01 ) ; C12N ( 86 ) PCT No .: PCT/ GB2016 /051116 2310 / 11 (2013 .01 ) $ 371 ( c ) ( 1 ) , ( 2 ) Date : Oct . 20 , 2017 (57 ) ABSTRACT The invention relates to oligonucleotides , e . g . , saRNAS Related U . S . Application Data useful in upregulating the expression of a target gene and (60 ) Provisional application No . 62 / 150 ,892 , filed on Apr. therapeutic compositions comprising such oligonucleotides . 22 , 2015 , provisional application No . 62 / 150 ,893 , Methods of using the oligonucleotides and the therapeutic filed on Apr. 22 , 2015 , provisional application No . compositions are also provided . 62 / 150 ,897 , filed on Apr. 22 , 2015 , provisional ap Specification includes a Sequence Listing . SARNA sense strand (Fessenger 3 ' SARNA antisense strand (Guide ) Mathew, Si Target antisense RNA transcript, e . g . NAT Target Coding strand Gene Transcription start site ( T55 ) TY{ { ? ? Targeted Target transcript , e . -

University of Copenhagen, Grønnegårdsvej 7, 1870 Frederiksberg, Denmark Cesses Covered Were Related to Each Other
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Copenhagen University Research Information System Copy number variations and genome-wide associations reveal putative genes and metabolic pathways involved with feed conversion ratio in beef cattle Satana, Miguel Henrique de Almedia ; Junior, Gerson Antônio Oliveira ; Cesar, Aline Silva Mello; Freua, Mateus Castelani; Gomes, Rodrigo da Costra ; Silva, Saulo da Luz e ; Leme, Paulo Roberto; Fukumasu, Heidge; Carvalho, Minos Esperandio; Ventura, Ricardo Vierira; Coutinho, Luiz Lehmann; Kadarmideen, Haja; Ferraz, José Bento Sterman Published in: Journal of Applied Genetics DOI: 10.1007/s13353-016-0344-7 Publication date: 2016 Document license: Other Citation for published version (APA): Satana, M. H. D. A., Junior, G. A. O., Cesar, A. S. M., Freua, M. C., Gomes, R. D. C., Silva, S. D. L. E., ... Ferraz, J. B. S. (2016). Copy number variations and genome-wide associations reveal putative genes and metabolic pathways involved with feed conversion ratio in beef cattle. Journal of Applied Genetics, 57(4), 495- 504. https://doi.org/10.1007/s13353-016-0344-7 Download date: 08. apr.. 2020 J Appl Genetics DOI 10.1007/s13353-016-0344-7 ANIMAL GENETICS • ORIGINAL PAPER Copy number variations and genome-wide associations reveal putative genes and metabolic pathways involved with the feed conversion ratio in beef cattle Miguel Henrique de Almeida Santana1,2 & Gerson Antônio Oliveira Junior3 & Aline Silva Mello Cesar3 & Mateus Castelani Freua2 & Rodrigo da Costa Gomes4 & Saulo da Luz e Silva2 & Paulo Roberto Leme2 & Heidge Fukumasu2 & Minos Esperândio Carvalho2 & Ricardo Vieira Ventura2,5 & Luiz Lehmann Coutinho6 & Haja N. -

Supplementary Data
1 SUPPLEMENTARY FILES – DESCRIPTION AND LEGENDS Supplementary Data: Validation of amplicons, analysis of grade III IDC-NST of indeterminate phenotype and quantification of PPM1D protein levels by densitometric analysis. Supplementary Figure 1: Validation of CCND1 and EGFR amplifications in a series of 91 grade III-IDC-NST and CCNE1 amplifications on selected cases. (A) i) H&E ii) CISH iii) IHC for a) luminal tumour for CCND1 showing amplification and protein over-expression, b) basal-like tumour for CCND1 with normal copy number and no protein expression, c) EGFR non-amplified tumour with no protein expression d) EGFR amplified tumour showing strong protein expression. Amplification of CCNE1 in basal-like breast cancers. (B) Genome plots of two cases exhibiting amplification with the smallest region of overlap highlighted (i). FISH confirmation of CCNE1 amplification with RP11-327I05 (CCNE1) (red), showing amplification > 5 copies per nucleus (Bii). Supplementary Figure 2: Microarray-based comparative genomic hybridisation analysis of five grade III invasive ductal carcinomas of no special type of indeterminate phenotype. A) Representative genome plots. Log2 ratios are plotted on the Y axis against each clone according to genomic location on the X axis. The centromere is represented by a vertical dotted line. BACs categorised as displaying genomic gains as defined by aws ratios > 0.08 are highlighted in green and those categorised as genomic losses as defined by aws ratios < -0.08 are highlighted in red. Bi) The proportion of tumours in which each clone is gained (green bars) or lost (red bars) is plotted (Y axis) for each BAC clone according to genomic location (X axis). -
Explorations in Olfactory Receptor Structure and Function by Jianghai
Explorations in Olfactory Receptor Structure and Function by Jianghai Ho Department of Neurobiology Duke University Date:_______________________ Approved: ___________________________ Hiroaki Matsunami, Supervisor ___________________________ Jorg Grandl, Chair ___________________________ Marc Caron ___________________________ Sid Simon ___________________________ [Committee Member Name] Dissertation submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy in the Department of Neurobiology in the Graduate School of Duke University 2014 ABSTRACT Explorations in Olfactory Receptor Structure and Function by Jianghai Ho Department of Neurobiology Duke University Date:_______________________ Approved: ___________________________ Hiroaki Matsunami, Supervisor ___________________________ Jorg Grandl, Chair ___________________________ Marc Caron ___________________________ Sid Simon ___________________________ [Committee Member Name] An abstract of a dissertation submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy in the Department of Neurobiology in the Graduate School of Duke University 2014 Copyright by Jianghai Ho 2014 Abstract Olfaction is one of the most primitive of our senses, and the olfactory receptors that mediate this very important chemical sense comprise the largest family of genes in the mammalian genome. It is therefore surprising that we understand so little of how olfactory receptors work. In particular we have a poor idea of what chemicals are detected by most of the olfactory receptors in the genome, and for those receptors which we have paired with ligands, we know relatively little about how the structure of these ligands can either activate or inhibit the activation of these receptors. Furthermore the large repertoire of olfactory receptors, which belong to the G protein coupled receptor (GPCR) superfamily, can serve as a model to contribute to our broader understanding of GPCR-ligand binding, especially since GPCRs are important pharmaceutical targets. -
The Spectra of Somatic Mutations Across Many Tumor Types
The spectra of somatic mutations across many tumor types Mike Lawrence Broad Institute of Harvard and MIT 1st Annual TCGA Scientific Symposium November 17, 2011 mutation rates across cancer Hematologic Carcinogens Childhood ?? ?? HPV & HPV mutation type C → T C → A C → G A → G A → T A → C OV mutation type C → T C → A C → G A → G A → T A → C mutation rate (per million sites) OV mutation type C → T C → A C → G A → G A → T A → C mutation rate (per million sites) GBM mutation type C → T C → A C → G A → G A → T A → C LUSC lung squamous mutation type C → T C → A C → G A → G A → T A → C LUAD lung adeno mutation type C → T C → A C → G A → G A → T A → C Melanoma mutation type C → T C → A C → G A → G A → T A → C cervical mutation type C → T C → A C → G A → G A → T A → C bladder total rate 100/Mb 10/Mb 1/Mb 0.1/Mb total rate type of spectrum Head&Neck HPV GBM HPV Bladder viral? Kidney Esophageal Colorectal Gastric Lung Melanoma GBM Kidney H&N Bladder Lung Gastric Colorectal Esophageal Melanoma finding significantly mutated genes patients tally significance MutSig scoring algorithm genes * patients tally significance MutSig scoring algorithm version 0 assume background mutation rate is: genes · uniform across sequence contexts · uniform across patients · uniform across genes * patients tally significance MutSig scoring algorithm version 1 assume background mutation rate is: genes · variable across sequence contexts · uniform across patients · uniform across genes * C→T (UV-induced) A→T patients tally significance MutSig scoring algorithm version