Carbohydrate Kinases: a Conserved Mechanism Across Differing Folds
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

METACYC ID Description A0AR23 GO:0004842 (Ubiquitin-Protein Ligase
Electronic Supplementary Material (ESI) for Integrative Biology This journal is © The Royal Society of Chemistry 2012 Heat Stress Responsive Zostera marina Genes, Southern Population (α=0. -

Molecular Characterization of Galactokinase Deficiency In
J Hum Genet (1999) 44:377–382 © Jpn Soc Hum Genet and Springer-Verlag 1999377 ORIGINAL ARTICLE Minoru Asada · Yoshiyuki Okano · Takuji Imamura Itsujin Suyama · Yutaka Hase · Gen Isshiki Molecular characterization of galactokinase deficiency in Japanese patients Received: May 19, 1999 / Accepted: August 21, 1999 Abstract Galactokinase (GALK) deficiency is an autoso- Key words Galactosemia · Galactokinase (GALK) · Muta- mal recessive disorder, which causes cataract formation in tion · Genotype · Phenotype children not maintained on a lactose-free diet. We charac- terized the human GALK gene by screening a Japanese genomic DNA phage library, and found that several nucle- otides in the 59-untranslated region and introns 1, 2, and 5 in Introduction our GALK genomic analysis differed from published data. A 20-bp tandem repeat was found in three places in intron Galactokinase (GALK: McKUSICK 230200) is the first 5, which were considered insertion sequences. We identified enzyme in the Leloir pathway of galactose metabolism; it five novel mutations in seven unrelated Japanese patients catalyzes the phosphorylation of galactose to galactose- with GALK deficiency. There were three missense muta- 1-phosphate. GALK deficiency, first described in 1965 tions and two deletions. All three missense mutations (Gitzelmann 1965), is an autosomal recessive genetic disor- (R256W, T344M, and G349S) occurred at CpG dinucle- der with an incidence of 1/1,000,000 in Japan (Aoki and otides, and the T344M and G349S mutations occurred in Wada 1988) on newborn mass screening and an incidence of the conserved region. The three missense mutations led to a 1/1,000,000 in Caucasians (Segal and Berry 1995). -

Gene Symbol Gene Description ACVR1B Activin a Receptor, Type IB
Table S1. Kinase clones included in human kinase cDNA library for yeast two-hybrid screening Gene Symbol Gene Description ACVR1B activin A receptor, type IB ADCK2 aarF domain containing kinase 2 ADCK4 aarF domain containing kinase 4 AGK multiple substrate lipid kinase;MULK AK1 adenylate kinase 1 AK3 adenylate kinase 3 like 1 AK3L1 adenylate kinase 3 ALDH18A1 aldehyde dehydrogenase 18 family, member A1;ALDH18A1 ALK anaplastic lymphoma kinase (Ki-1) ALPK1 alpha-kinase 1 ALPK2 alpha-kinase 2 AMHR2 anti-Mullerian hormone receptor, type II ARAF v-raf murine sarcoma 3611 viral oncogene homolog 1 ARSG arylsulfatase G;ARSG AURKB aurora kinase B AURKC aurora kinase C BCKDK branched chain alpha-ketoacid dehydrogenase kinase BMPR1A bone morphogenetic protein receptor, type IA BMPR2 bone morphogenetic protein receptor, type II (serine/threonine kinase) BRAF v-raf murine sarcoma viral oncogene homolog B1 BRD3 bromodomain containing 3 BRD4 bromodomain containing 4 BTK Bruton agammaglobulinemia tyrosine kinase BUB1 BUB1 budding uninhibited by benzimidazoles 1 homolog (yeast) BUB1B BUB1 budding uninhibited by benzimidazoles 1 homolog beta (yeast) C9orf98 chromosome 9 open reading frame 98;C9orf98 CABC1 chaperone, ABC1 activity of bc1 complex like (S. pombe) CALM1 calmodulin 1 (phosphorylase kinase, delta) CALM2 calmodulin 2 (phosphorylase kinase, delta) CALM3 calmodulin 3 (phosphorylase kinase, delta) CAMK1 calcium/calmodulin-dependent protein kinase I CAMK2A calcium/calmodulin-dependent protein kinase (CaM kinase) II alpha CAMK2B calcium/calmodulin-dependent -

In the Field of Clinical Examination, the Measurement of Creatine
J. Clin. Biochem. Nutr., 3, 17-25, 1987 Bacterial Glucokinase as an Enzymic Reagent of Good Stability for Measurement of Creatine Kinase Activity Hitoshi KONDO, * Takanari SHIRAISHI, Masao KAGEYAMA, Kazuhiko NAGATA, and Kosuke TOMITA Research and Development Center, UNITIKA Ltd., Uji 611, Japan (Received January 10, 1987) Summary An enzymic reagent, that has long-term stability even in the liquid state, was successfully employed for the measurement of serum creatine kinase (CK, EC 2.7.3.2) activity. The enzyme used was the thermostable glucokinase (GlcK, EC 2.7.1.2) obtained from the thermo- phile Bacillus stearothermophilus. The reagent was found to be stable in solution for about one month at 6•Ž and for about one week at 30•Ž. This substitution of glucokinase for the hexokinase of the most commonly used hexokinase-glucose-6-phosphate dehydrogenase (HK-G6PDH) method results in a remarkable improvement of the method. The CK activity measured by the GlcK-G6PDH method was linear up to about 2,000 U/ liter at 37•Ž. The GlcK-G6PDH method was found to give a satisfactory precision and reproducibility (coefficient of variation less than 2.17%). Over a wide range of CK activity, an excellent agreement was obtained between the GlcK-G6PDH and the HK-G6PDH methods. Furthermore several coexistents and anticoagulants were found to have little effect on the measured value of CK activity by the GlcK-G6PDH method. Key Words: creatine kinase activity, glucokinase, improved stability of reagent, creatine kinase determination, thermostable enzyme In the field of clinical examination, the measurement of creatine kinase (CK, ATP : creatine phosphotransferase, EC 2.7.3.2) activity in serum is one of the important examinations usually employed for diagnosis of cardiac diseases such as myocardial infarction or muscular diseases such as progressive muscular dystrophy. -

A Disease Spectrum for ITPA Variation: Advances in Biochemical and Clinical Research Nicholas E
Burgis Journal of Biomedical Science (2016) 23:73 DOI 10.1186/s12929-016-0291-y REVIEW Open Access A disease spectrum for ITPA variation: advances in biochemical and clinical research Nicholas E. Burgis Abstract Human ITPase (encoded by the ITPA gene) is a protective enzyme which acts to exclude noncanonical (deoxy) nucleoside triphosphates ((d)NTPs) such as (deoxy)inosine 5′-triphosphate ((d)ITP), from (d)NTP pools. Until the last few years, the importance of ITPase in human health and disease has been enigmatic. In 2009, an article was published demonstrating that ITPase deficiency in mice is lethal. All homozygous null offspring died before weaning as a result of cardiomyopathy due to a defect in the maintenance of quality ATP pools. More recently, a whole exome sequencing project revealed that very rare, severe human ITPA mutation results in early infantile encephalopathy and death. It has been estimated that nearly one third of the human population has an ITPA status which is associated with decreased ITPase activity. ITPA status has been linked to altered outcomes for patients undergoing thiopurine or ribavirin therapy. Thiopurine therapy can be toxic for patients with ITPA polymorphism, however, ITPA polymorphism is associated with improved outcomes for patients undergoing ribavirin treatment. ITPA polymorphism has also been linked to early-onset tuberculosis susceptibility. These data suggest a spectrum of ITPA-related disease exists in human populations. Potentially, ITPA status may affect a large number of patient outcomes, suggesting that modulation of ITPase activity is an important emerging avenue for reducing the number of negative outcomes for ITPA-related disease. -

Indications for a Central Role of Hexokinase Activity in Natural Variation of Heat Acclimation in Arabidopsis Thaliana
Preprints (www.preprints.org) | NOT PEER-REVIEWED | Posted: 14 June 2020 doi:10.20944/preprints202006.0169.v1 Article Indications for a central role of hexokinase activity in natural variation of heat acclimation in Arabidopsis thaliana Vasil Atanasov §, Lisa Fürtauer § and Thomas Nägele * LMU Munich, Plant Evolutionary Cell Biology, Großhaderner Str. 2-4, 82152 Planegg, Germany § Authors contributed equally * Correspondence: [email protected] Abstract: Diurnal and seasonal changes of abiotic environmental factors shape plant performance and distribution. Changes of growth temperature and light intensity may vary significantly on a diurnal, but also on a weekly or seasonal scale. Hence, acclimation to a changing temperature and light regime is essential for plant survival and propagation. In the present study, we analyzed photosynthetic CO2 assimilation and metabolic regulation of the central carbohydrate metabolism in two natural accessions of Arabidopsis thaliana originating from Russia and south Italy during exposure to heat and a combination of heat and high light. Our findings indicate that it is hardly possible to predict photosynthetic capacities to fix CO2 under combined stress from single stress experiments. Further, capacities of hexose phosphorylation were found to be significantly lower in the Italian than in the Russian accession which could explain an inverted sucrose-to-hexose ratio. Together with the finding of significantly stronger accumulation of anthocyanins under heat/high light these observations indicate a central role of hexokinase activity in stabilization of photosynthetic capacities within a changing environment. Keywords: photosynthesis; carbohydrate metabolism; hexokinase; heat acclimation; environmental changes; natural variation; high light; combined stress. 1. Introduction Changes of growth temperature and light intensity broadly affect plant molecular, physiological and developmental processes. -

Non-Homologous Isofunctional Enzymes: a Systematic Analysis Of
Omelchenko et al. Biology Direct 2010, 5:31 http://www.biology-direct.com/content/5/1/31 RESEARCH Open Access Non-homologousResearch isofunctional enzymes: A systematic analysis of alternative solutions in enzyme evolution Marina V Omelchenko, Michael Y Galperin*, Yuri I Wolf and Eugene V Koonin Abstract Background: Evolutionarily unrelated proteins that catalyze the same biochemical reactions are often referred to as analogous - as opposed to homologous - enzymes. The existence of numerous alternative, non-homologous enzyme isoforms presents an interesting evolutionary problem; it also complicates genome-based reconstruction of the metabolic pathways in a variety of organisms. In 1998, a systematic search for analogous enzymes resulted in the identification of 105 Enzyme Commission (EC) numbers that included two or more proteins without detectable sequence similarity to each other, including 34 EC nodes where proteins were known (or predicted) to have distinct structural folds, indicating independent evolutionary origins. In the past 12 years, many putative non-homologous isofunctional enzymes were identified in newly sequenced genomes. In addition, efforts in structural genomics resulted in a vastly improved structural coverage of proteomes, providing for definitive assessment of (non)homologous relationships between proteins. Results: We report the results of a comprehensive search for non-homologous isofunctional enzymes (NISE) that yielded 185 EC nodes with two or more experimentally characterized - or predicted - structurally unrelated proteins. Of these NISE sets, only 74 were from the original 1998 list. Structural assignments of the NISE show over-representation of proteins with the TIM barrel fold and the nucleotide-binding Rossmann fold. From the functional perspective, the set of NISE is enriched in hydrolases, particularly carbohydrate hydrolases, and in enzymes involved in defense against oxidative stress. -

Labeled in Thecourse of Glycolysis, Since Phosphoglycerate Kinase
THE STATE OF MAGNESIUM IN CELLS AS ESTIMATED FROM THE ADENYLATE KINASE EQUILIBRIUM* BY TRWIN A. RoSE THE INSTITUTE FOR CANCER RESEARCH, PHILADELPHIA Communicated by Thomas F. Anderson, August 30, 1968 Magnesium functions in many enzymatic reactions as a cofactor and in com- plex with nucleotides acting as substrates. Numerous examples of a possible regulatory role of Mg can be cited from studies with isolated enzymes,'- and it is known that Mg affects the structural integrity of macromolecules such as trans- fer RNA" and functional elements such as ribosomes.'0 The major problem in translating this information on isolated preparations to the functioning cell is the difficulty in determining the distribution of Mg and the nucleotides among the free and complexed forms that function in the region of the cell for which this information is desired. Nanningall based an attempt to calculate the free Mg2+ and Ca2+ ion concentrations of frog muscle on the total content of these metals and of the principal known ligands (adenosine 5'-triphosphate (ATP), creatine-P, and myosin) and the dissociation constants of the complexes. However, this method suffers from the necessity of evaluating the contribution of all ligands as well as from the assumption that all the known ligands are contributing their full complexing capacity. During studies concerned with the control of glycolysis in red cells and the control of the phosphoglycerate kinase step in particular, it became important to determine the fractions of the cell's ATP and adenosine 5'-diphosphate (ADP) that were present as Mg complexes. Just as the problem of determining the distribution of protonated and dissociated forms of an acid can be solved from a knowledge of pH and pKa of the acid, so it would be possible to determine the liganded and free forms of all rapidly established Mg complexes from a knowledge of Mg2+ ion concentration and the appropriate dissociation constants. -

Adaptive Laboratory Evolution Enhances Methanol Tolerance and Conversion in Engineered Corynebacterium Glutamicum
ARTICLE https://doi.org/10.1038/s42003-020-0954-9 OPEN Adaptive laboratory evolution enhances methanol tolerance and conversion in engineered Corynebacterium glutamicum Yu Wang 1, Liwen Fan1,2, Philibert Tuyishime1, Jiao Liu1, Kun Zhang1,3, Ning Gao1,3, Zhihui Zhang1,3, ✉ ✉ 1234567890():,; Xiaomeng Ni1, Jinhui Feng1, Qianqian Yuan1, Hongwu Ma1, Ping Zheng1,2,3 , Jibin Sun1,3 & Yanhe Ma1 Synthetic methylotrophy has recently been intensively studied to achieve methanol-based biomanufacturing of fuels and chemicals. However, attempts to engineer platform micro- organisms to utilize methanol mainly focus on enzyme and pathway engineering. Herein, we enhanced methanol bioconversion of synthetic methylotrophs by improving cellular tolerance to methanol. A previously engineered methanol-dependent Corynebacterium glutamicum is subjected to adaptive laboratory evolution with elevated methanol content. Unexpectedly, the evolved strain not only tolerates higher concentrations of methanol but also shows improved growth and methanol utilization. Transcriptome analysis suggests increased methanol con- centrations rebalance methylotrophic metabolism by down-regulating glycolysis and up- regulating amino acid biosynthesis, oxidative phosphorylation, ribosome biosynthesis, and parts of TCA cycle. Mutations in the O-acetyl-L-homoserine sulfhydrylase Cgl0653 catalyzing formation of L-methionine analog from methanol and methanol-induced membrane-bound transporter Cgl0833 are proven crucial for methanol tolerance. This study demonstrates the importance of -
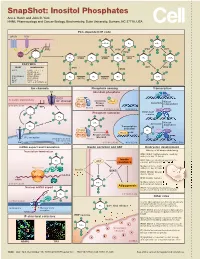
Snapshot: Inositol Phosphates Ace J
SnapShot: Inositol Phosphates Ace J. Hatch and John D. York HHMI, Pharmacology and Cancer Biology, Biochemistry, Duke University, Durham, NC 27710, USA PLC-dependent IP code GPCR RTK O O O O O O 5-PP-IP4 IP4 5-IP7 O O O O O O PIP2 O IP6K O IP6K O VIP1 O O O 2 O ITPK1 O 13 O PLC 2 O O O O O O O O 4 6 13 O 5 IP3 IPMK IP4 IPMK IP5 IPK1 IP6 1,5-IP8 4 6 O O 5 O O O O O O O O ENZYMES O O O O O O YEAST MAMMALIAN IP3K VIP1 IP6K IPMK PLC1 PLCβ, γ, δ, ε, ζ, η O - IP3KA, B, C - ITPK1 (IP56K) O O O O O O O O IPK2(ARG82) IPMK (IPK2) IP4 IP3 IP4 1-IP7 IPK1 IPK1 (IP5K) INPP5 ITPK1 KCS1 IP6K1, 2, 3 O O O O O O VIP1 VIP1, 2 (PPIP5K1, 2) O O Ion channels Phosphate sensing Transcription Cl- Abundant phosphate MCM1 ARG80 CIC3 P PLASMA MEMBRANE - Pho80 Cl channel Pho4 Kinase Kinase Assembly Pho85 independent CYTOPLASM activity 2 O PIP2 Pho81 13 CYTOPLASM NUCLEUS IPK2 ARG81 4 6 Phosphate starvation MCM1-ArgR O 5 O complex O O IP4 O O O O O O O 1-IP7 Kinase Activation dependent IP3 O O Transcription O O O activated Pho80 IP4 O X Pho4 O O Pho85 Kinase activity IP receptor blocked O 3 ENDOPLASMIC Pho81 RETICULUM Ca2+ CYTOPLASM NUCLEUS NUCLEUS mRNA export and translation Insulin secretion and AKT Embryonic development Translation termination Effects of IP kinase deficiency O IPMK (IPK2): Multiple defects, death by embryonic day 10 (mice) O O Insulin IPK1: Cillia are shortened and immotile IP6 AKT resistance causing patterning defects (zebrash) O O Multiple defects, death by Ribosome O embryonic day 8.5 (mice) GleI eRF1 Insulin GSK3β Dbp5 ITPK1 (IP56K): Neural tube -

(LRV1) Pathogenicity Factor
Antiviral screening identifies adenosine analogs PNAS PLUS targeting the endogenous dsRNA Leishmania RNA virus 1 (LRV1) pathogenicity factor F. Matthew Kuhlmanna,b, John I. Robinsona, Gregory R. Bluemlingc, Catherine Ronetd, Nicolas Faseld, and Stephen M. Beverleya,1 aDepartment of Molecular Microbiology, Washington University School of Medicine in St. Louis, St. Louis, MO 63110; bDepartment of Medicine, Division of Infectious Diseases, Washington University School of Medicine in St. Louis, St. Louis, MO 63110; cEmory Institute for Drug Development, Emory University, Atlanta, GA 30329; and dDepartment of Biochemistry, University of Lausanne, 1066 Lausanne, Switzerland Contributed by Stephen M. Beverley, December 19, 2016 (sent for review November 21, 2016; reviewed by Buddy Ullman and C. C. Wang) + + The endogenous double-stranded RNA (dsRNA) virus Leishmaniavirus macrophages infected in vitro with LRV1 L. guyanensis or LRV2 (LRV1) has been implicated as a pathogenicity factor for leishmaniasis Leishmania aethiopica release higher levels of cytokines, which are in rodent models and human disease, and associated with drug-treat- dependent on Toll-like receptor 3 (7, 10). Recently, methods for ment failures in Leishmania braziliensis and Leishmania guyanensis systematically eliminating LRV1 by RNA interference have been − infections. Thus, methods targeting LRV1 could have therapeutic ben- developed, enabling the generation of isogenic LRV1 lines and efit. Here we screened a panel of antivirals for parasite and LRV1 allowing the extension of the LRV1-dependent virulence paradigm inhibition, focusing on nucleoside analogs to capitalize on the highly to L. braziliensis (12). active salvage pathways of Leishmania, which are purine auxo- A key question is the relevancy of the studies carried out in trophs. -

(12) Patent Application Publication (10) Pub. No.: US 2014/0155567 A1 Burk Et Al
US 2014O155567A1 (19) United States (12) Patent Application Publication (10) Pub. No.: US 2014/0155567 A1 Burk et al. (43) Pub. Date: Jun. 5, 2014 (54) MICROORGANISMS AND METHODS FOR (60) Provisional application No. 61/331,812, filed on May THE BIOSYNTHESIS OF BUTADENE 5, 2010. (71) Applicant: Genomatica, Inc., San Diego, CA (US) Publication Classification (72) Inventors: Mark J. Burk, San Diego, CA (US); (51) Int. Cl. Anthony P. Burgard, Bellefonte, PA CI2P 5/02 (2006.01) (US); Jun Sun, San Diego, CA (US); CSF 36/06 (2006.01) Robin E. Osterhout, San Diego, CA CD7C II/6 (2006.01) (US); Priti Pharkya, San Diego, CA (52) U.S. Cl. (US) CPC ................. CI2P5/026 (2013.01); C07C II/I6 (2013.01); C08F 136/06 (2013.01) (73) Assignee: Genomatica, Inc., San Diego, CA (US) USPC ... 526/335; 435/252.3:435/167; 435/254.2: (21) Appl. No.: 14/059,131 435/254.11: 435/252.33: 435/254.21:585/16 (22) Filed: Oct. 21, 2013 (57) ABSTRACT O O The invention provides non-naturally occurring microbial Related U.S. Application Data organisms having a butadiene pathway. The invention addi (63) Continuation of application No. 13/101,046, filed on tionally provides methods of using Such organisms to produce May 4, 2011, now Pat. No. 8,580,543. butadiene. Patent Application Publication Jun. 5, 2014 Sheet 1 of 4 US 2014/O155567 A1 ?ueudos!SMS |?un61– Patent Application Publication Jun. 5, 2014 Sheet 2 of 4 US 2014/O155567 A1 VOJ OO O Z?un61– Patent Application Publication US 2014/O155567 A1 {}}} Hººso Patent Application Publication Jun.