Spatio-Temporal Expression Profile of Sirtuins During Aging of the Annual
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Deterministic Shifts in Molecular Evolution Correlate with Convergence to Annualism in Killifishes
bioRxiv preprint doi: https://doi.org/10.1101/2021.08.09.455723; this version posted August 10, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Deterministic shifts in molecular evolution correlate with convergence to annualism in killifishes Andrew W. Thompson1,2, Amanda C. Black3, Yu Huang4,5,6 Qiong Shi4,5 Andrew I. Furness7, Ingo, Braasch1,2, Federico G. Hoffmann3, and Guillermo Ortí6 1Department of Integrative Biology, Michigan State University, East Lansing, Michigan 48823, USA. 2Ecology, Evolution & Behavior Program, Michigan State University, East Lansing, MI, USA. 3Department of Biochemistry, Molecular Biology, Entomology, & Plant Pathology, Mississippi State University, Starkville, MS 39759, USA. 4Shenzhen Key Lab of Marine Genomics, Guangdong Provincial Key Lab of Molecular Breeding in Marine Economic Animals, BGI Marine, Shenzhen 518083, China. 5BGI Education Center, University of Chinese Academy of Sciences, Shenzhen 518083, China. 6Department of Biological Sciences, The George Washington University, Washington, DC 20052, USA. 7Department of Biological and Marine Sciences, University of Hull, UK. Corresponding author: Andrew W. Thompson, [email protected] bioRxiv preprint doi: https://doi.org/10.1101/2021.08.09.455723; this version posted August 10, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Abstract: The repeated evolution of novel life histories correlating with ecological variables offer opportunities to test scenarios of convergence and determinism in genetic, developmental, and metabolic features. Here we leverage the diversity of aplocheiloid killifishes, a clade of teleost fishes that contains over 750 species on three continents. -

Nothobranchius As a Model for Aging Studies. a Review
Volume 5, Number 2; xxx-xx, April 2014 http://dx.doi.org/10.14336/AD Review Article Nothobranchius as a model for aging studies. A review Alejandro Lucas-Sánchez 1, *, Pedro Francisco Almaida-Pagán 2, Pilar Mendiola 1, Jorge de Costa 1 1 Department of Physiology. Faculty of Biology. University of Murcia. 30100 Murcia, Spain. 2 Institute of Aquaculture, School of Natural Sciences, University of Stirling, Stirling FK9 4LA, UK. [Received October 17, 2013; Revised December 4, 2013; Accepted December 4, 2013] ABSTRACT: In recent decades, the increase in human longevity has made it increasingly important to expand our knowledge on aging. To accomplish this, the use of animal models is essential, with the most common being mouse (phylogenetically similar to humans, and a model with a long life expectancy) and Caenorhabditis elegans (an invertebrate with a short life span, but quite removed from us in evolutionary terms). However, some sort of model is needed to bridge the differences between those mentioned above, achieving a balance between phylogenetic distance and life span. Fish of the genus Nothobranchius were suggested 10 years ago as a possible alternative for the study of the aging process. In the meantime, numerous studies have been conducted at different levels: behavioral (including the study of the rest-activity rhythm), populational, histochemical, biochemical and genetic, among others, with very positive results. This review compiles what we know about Nothobranchius to date, and examines its future prospects as a true alternative to the classic models for studies on aging. Key words: Aging, Fish, killifish, Nothobranchius Aging is a multifactorial process that involves numerous To be considered a model for aging, an animal must mechanisms that operate during the normal life cycle of have a number of features. -
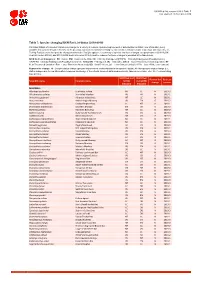
Table 7: Species Changing IUCN Red List Status (2018-2019)
IUCN Red List version 2019-3: Table 7 Last Updated: 10 December 2019 Table 7: Species changing IUCN Red List Status (2018-2019) Published listings of a species' status may change for a variety of reasons (genuine improvement or deterioration in status; new information being available that was not known at the time of the previous assessment; taxonomic changes; corrections to mistakes made in previous assessments, etc. To help Red List users interpret the changes between the Red List updates, a summary of species that have changed category between 2018 (IUCN Red List version 2018-2) and 2019 (IUCN Red List version 2019-3) and the reasons for these changes is provided in the table below. IUCN Red List Categories: EX - Extinct, EW - Extinct in the Wild, CR - Critically Endangered [CR(PE) - Critically Endangered (Possibly Extinct), CR(PEW) - Critically Endangered (Possibly Extinct in the Wild)], EN - Endangered, VU - Vulnerable, LR/cd - Lower Risk/conservation dependent, NT - Near Threatened (includes LR/nt - Lower Risk/near threatened), DD - Data Deficient, LC - Least Concern (includes LR/lc - Lower Risk, least concern). Reasons for change: G - Genuine status change (genuine improvement or deterioration in the species' status); N - Non-genuine status change (i.e., status changes due to new information, improved knowledge of the criteria, incorrect data used previously, taxonomic revision, etc.); E - Previous listing was an Error. IUCN Red List IUCN Red Reason for Red List Scientific name Common name (2018) List (2019) change version Category -

The Neotropical Genus Austrolebias: an Emerging Model of Annual Killifishes Nibia Berois1, Maria J
lopmen ve ta e l B D io & l l o l g e y C Cell & Developmental Biology Berois, et al., Cell Dev Biol 2014, 3:2 ISSN: 2168-9296 DOI: 10.4172/2168-9296.1000136 Review Article Open Access The Neotropical Genus Austrolebias: An Emerging Model of Annual Killifishes Nibia Berois1, Maria J. Arezo1 and Rafael O. de Sá2* 1Departamento de Biologia Celular y Molecular, Facultad de Ciencias, Universidad de la República, Montevideo, Uruguay 2Department of Biology, University of Richmond, Richmond, Virginia, USA *Corresponding author: Rafael O. de Sá, Department of Biology, University of Richmond, Richmond, Virginia, USA, Tel: 804-2898542; Fax: 804-289-8233; E-mail: [email protected] Rec date: Apr 17, 2014; Acc date: May 24, 2014; Pub date: May 27, 2014 Copyright: © 2014 Rafael O. de Sá, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. Abstract Annual fishes are found in both Africa and South America occupying ephemeral ponds that dried seasonally. Neotropical annual fishes are members of the family Rivulidae that consist of both annual and non-annual fishes. Annual species are characterized by a prolonged embryonic development and a relatively short adult life. Males and females show striking sexual dimorphisms, complex courtship, and mating behaviors. The prolonged embryonic stage has several traits including embryos that are resistant to desiccation and undergo up to three reversible developmental arrests until hatching. These unique developmental adaptations are closely related to the annual fish life cycle and are the key to the survival of the species. -

Draft Risk Assessment Report Nothobranchius Furzeri (Turquoise
APPLICATION TO AMEND THE LIST OF SPECIMENS SUITABLE FOR LIVE IMPORT Draft Risk Assessment Report 1. Taxonomy of the species a. Family name: Aplocheilidae b. Genus name: Nothobranchius c. Species: N. furzeri d. Subspecies: No e. Taxonomic Reference: http://www.uniprot.org/taxonomy/105023 f. Common Names: Turquoise killifish g. Is the species a genetically-modified organism (GMO)? No 2. Status of the species under CITES Species is not listed in the CITES Appendices. 3. Ecology of the species a. Longevity: what is the average lifespan of the species in the wild and in captivity? The average lifespan of the species in captivity is 13 weeks (1). Wild derived N. furzeri from three different habitats showed a maximum lifespan of 25-32 weeks (1). b. What is the maximum length and weight that the species attains? Provide information on the size and weight range for males and females of the species. As per the original formal description of the species, the standard length of N. furzeri from a wild population ranged from 2.5cm to 4.4cm (2). The maximum length the species can attain is 7cm (3). The mean maximal body weight reported for males is 3.8 ± 1.1 g (range: 0.5 – 6.7 g) and 2.2 ± 0.6 g (range: 0.2 - 3.6 g) for females (4). Growth rates of Nothobranchius in the wild have been reported to be similar to those in captivity (5). c. Discuss the identification of the individuals in this species, including if the sexes of the species are readily distinguishable, and if the species is difficult to distinguish from other species. -

Miombo Ecoregion Vision Report
MIOMBO ECOREGION VISION REPORT Jonathan Timberlake & Emmanuel Chidumayo December 2001 (published 2011) Occasional Publications in Biodiversity No. 20 WWF - SARPO MIOMBO ECOREGION VISION REPORT 2001 (revised August 2011) by Jonathan Timberlake & Emmanuel Chidumayo Occasional Publications in Biodiversity No. 20 Biodiversity Foundation for Africa P.O. Box FM730, Famona, Bulawayo, Zimbabwe PREFACE The Miombo Ecoregion Vision Report was commissioned in 2001 by the Southern Africa Regional Programme Office of the World Wide Fund for Nature (WWF SARPO). It represented the culmination of an ecoregion reconnaissance process led by Bruce Byers (see Byers 2001a, 2001b), followed by an ecoregion-scale mapping process of taxa and areas of interest or importance for various ecological and bio-physical parameters. The report was then used as a basis for more detailed discussions during a series of national workshops held across the region in the early part of 2002. The main purpose of the reconnaissance and visioning process was to initially outline the bio-physical extent and properties of the so-called Miombo Ecoregion (in practice, a collection of smaller previously described ecoregions), to identify the main areas of potential conservation interest and to identify appropriate activities and areas for conservation action. The outline and some features of the Miombo Ecoregion (later termed the Miombo– Mopane Ecoregion by Conservation International, or the Miombo–Mopane Woodlands and Grasslands) are often mentioned (e.g. Burgess et al. 2004). However, apart from two booklets (WWF SARPO 2001, 2003), few details or justifications are publically available, although a modified outline can be found in Frost, Timberlake & Chidumayo (2002). Over the years numerous requests have been made to use and refer to the original document and maps, which had only very restricted distribution. -

33723007.Pdf
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Aquatic Commons SPONSORS Hydrobiologists from East, Central and West Africa with substantial support from other African countries, Fishery Scientists in the United States, Canada, U.K., Europe, Soviet Union and Israel. OBSERVATIONS EDITOR SIZE ON BOTTO Dr. J. Okedi, Director, E.A.F.F.R.O., Jinja, Uganda. WITH EMPHASI: EDITORIAL BOARD Mr. M. Abolarin, Co-Manager, Kainji Lake Professor W.B. Banage, Makelere Univer Project, Lagos, Nigeria. sity, Kampala, Uganda. ALMO J. CORDONE Al UNDP(SF)/ FAO Lake Vici Mr. J. Kambona, Chief Fisheries Officer, Mr. R.E. Morris, Director, E.A.M.F.R.O., Freshwater Fisheries Resel Dar es Salaam, Tanzania. Zanzibar. Mr. J. Mubanga, Director, Fisheries Division, Dr. T. Petr, Senior Lecturer in Hydrobiology, Chilanga, Zambia.. Makerere University, Kaflpala, Uganda. Dr. L. Obeng, Director, Institute of Aquatic Professor Mohamed Hyder, University of Biology, Achimota, Ghana. Nairobi, Kenya. Mr. N. Odero, Director ofFisheries, Nairobi, Professor, A. F. De Bont, Universite de Kenya. Kinshasa, Kinshasa XI, Republique Demo Mr. S.N. Semakula, Chief Fisheries Officer, cratique du Zaire Entebbe, Uganda. PROGRAMME The African Jout-nal of Tropical Hydrobiology and Fisheries will only accept original and well supported ideas on techniques, methodology and research findings from scientists, fishery officers,. fishery economists and sociologists. The Journal will therefore strengthen the African lesearch scientist by making research material available and also increasing the awareness and utility of aquatic resources. Its quality will conform to International standards, and will be published in English and French. -

The Evolutionary Ecology of African Annual Fishes
CHAPTER 9 The Evolutionary Ecology of African Annual Fishes Martin Reichard CONTENTS 9.1 Distribution and Biogeography ............................................................................................. 133 9.1.1 Habitat Types ............................................................................................................ 134 9.1.2 Species Distribution and Range Size ........................................................................ 136 9.1.3 Climatic Conditions .................................................................................................. 136 9.1.4 Biogeography ............................................................................................................ 139 9.1.5 Dispersal and Colonization ....................................................................................... 140 9.2 Species Coexistence .............................................................................................................. 142 9.2.1 Community Assembly .............................................................................................. 142 9.2.2 Habitat Use ............................................................................................................... 144 9.2.3 Morphology and Diet ................................................................................................ 144 9.3 Population Ecology ............................................................................................................... 145 9.3.1 Population Genetic Structure ................................................................................... -

Cyprinodontiformes, Nothobranchiidae)
COMPARATIVE A peer-reviewed open-access journal CompCytogen 10(3): Divergent439–445 (2016) karyotypes of the annual killifish genusNothobranchius ... 439 doi: 10.3897/CompCytogen.v10i3.9863 SHORT COMMUNICATION Cytogenetics http://compcytogen.pensoft.net International Journal of Plant & Animal Cytogenetics, Karyosystematics, and Molecular Systematics Divergent karyotypes of the annual killifish genus Nothobranchius (Cyprinodontiformes, Nothobranchiidae) Eugene Krysanov1, Tatiana Demidova1, Bela Nagy2 1 Severtsov Institute of Ecology and Evolution, Russian Academy of Sciences, Leninsky prospect, Moscow, 119071 Russia 2 30, rue du Mont Ussy, 77300 Fontainebleau, France Corresponding author: Eugene Krysanov ([email protected]) Academic editor: I. Kuznetsova | Received 13 July 2016 | Accepted 18 August 2016 | Published 16 September 2016 http://zoobank.org/516FBC12-825D-46E0-BAAA-25ABA00F8607 Citation: Krysanov E, Demidova T, Nagy B (2016) Divergent karyotypes of the annual killifish genus Nothobranchius (Cyprinodontiformes; Nothobranchiidae). Comparative Cytogenetics 10(3): 439–445. doi: 10.3897/CompCytogen. v10i3.9863 Abstract Karyotypes of two species of the African annual killifish genus Nothobranchius Peters, 1868, N. brieni Poll, 1938 and Nothobranchius sp. from Kasenga (D.R. Congo) are described. Both species displayed dip- loid chromosome number 2n = 49/50 for males and females respectively with multiple-sex chromosome system type X1X2Y/X1X1X2X2. The karyotypes of studied species are considerably different from those previously reported for the genus Nothobranchius and similar to the Actinopterygii conservative karyotype. Keywords Africa, chromosome number, karyotype, killifish, Nothobranchius Introduction Annual killifishes belonging to the genusNothobranchius Peters, 1868 are mainly dis- tributed in eastern Africa but several species are found in central Africa (Wildekamp 2004). They inhabit temporary pools that dry out during the dry season and have spe- cific adaptations for extreme environments. -

Nothobranchius Nubaensis (Cyprinodontiformes: Nothobranchiidae) a New Annual Killifish from Sudan and Ethiopia
aqua, International Journal of Ichthyology Nothobranchius nubaensis (Cyprinodontiformes: Nothobranchiidae) a new annual killifish from Sudan and Ethiopia Stefano Valdesalici 1, Marc Bellemans 2, Kiril Kardashev 3, Alexander Golubtsov 4 1) Via Cà Bertacchi 5, 42030 VianO (RE), Italy. E-mail: [email protected] and [email protected] 2) MOrtselsesteenweg 138, 2540 HOve, Belgium 3) Stramna 4, 2600 Dupnitsa, Bulgaria 4) LabOratOry Of LOwer Vertebrate EcOlOgy, A.N. SevertsOv Institute Of EcOlOgy & EvOlutiOn, Russian Academy Of Sciences, Leninskii Pr. 33, MOscOw119071, Russia Received: 19 March 2009 – Accepted: 04 July 2009 Abstract Résumé A new species Of annual killifish, Nothobranchius nubaensis , Une nOuvelle espèce de killy annuel, Nothobranchius is described based On specimens cOllected frOm ephemeral nubaensis, est décrite sur base de spécimens cOllectés dans water bOdies in central Sudan and sOuth -west EthiOpia. The des pOches d’eau tempOraires du centre du SOudan et du new species is distinguished frOm the Other members Of the sud-Ouest de l’EthiOpie. La nOuvelle espèce se distingue des Nothobranchius ugandensis species grOup by the fOllOwing autres membres du grOupe Nothobranchius ugandensis par cOmbinatiOn Of characters : 17-19 dOrsal fin rays; 17-19 anal la cOmbinaisOn des caractéristiques suivantes: 17 à 19 fin rays; 29-30 scales in median lateral series; dOrsal and anal rayOns dOrsaux; 17 à 19 rayOns à l’anale, 29 à 30 écailles fins in males with shOrt filamentOus rays; pelvic fins shOrt, dans les rangées latérales médianes; -

Trophic Niche Partitioning in Communities of African Annual Fish
Hydrobiologia (2014) 721:99–106 DOI 10.1007/s10750-013-1652-0 PRIMARY RESEARCH PAPER Trophic niche partitioning in communities of African annual fish: evidence from stable isotopes Matej Polacˇik • Chris Harrod • Radim Blazˇek • Martin Reichard Received: 6 March 2013 / Revised: 25 July 2013 / Accepted: 3 August 2013 / Published online: 17 August 2013 Ó Springer Science+Business Media Dordrecht 2013 Abstract Annual killifish of the genus Nothobranch- separated the trophic niche and trophic position of N. ius often co-occur in temporary savannah pools. Their pienaari but failed to find any difference between N. space- and time-limited environment does not allow furzeri/N. kadleci and N. orthonotus. No segregation for any substantial habitat or temporal segregation. was found at the sites with low prey diversity. In Coexisting species are therefore predicted to have well contrast, SCA identified N. orthonotus as the species separated trophic niches to avoid intense food compe- with the most distinct trophic niche. We discuss the tition. Although in a previous ‘‘snapshot’’ study using effect of prey diversity and different sensitivities of stomach content analysis (SCA), the trophic niches of stomach content and stable isotope analysis in general three sympatric species (N. furzeri, N. orthonotus, and and conclude that the trophic niches of the three N. pienaari) were found to vary among species, the sympatric Nothobranchius species are well separated. difference was relatively weak and inconsistent across different sites. Here, we used the time-integrative Keywords Nothobranchius Á Coexistence Á capacity of stable isotope analysis to test whether the Niche separation Á Sympatric Á Extreme trophic niches of sympatric Mozambican Notho- environment Á Africa branchius are more distinct over a long-term period. -

Strong Population Genetic Structuring in an Annual Fish, Nothobranchius
Bartáková et al. BMC Evolutionary Biology 2013, 13:196 http://www.biomedcentral.com/1471-2148/13/196 RESEARCH ARTICLE Open Access Strong population genetic structuring in an annual fish, Nothobranchius furzeri, suggests multiple savannah refugia in southern Mozambique Veronika Bartáková1,2, Martin Reichard1, Karel Janko1,3, Matej Polačik1, Radim Blažek1, Kathrin Reichwald4, Alessandro Cellerino4,5 and Josef Bryja1,2* Abstract Background: Intraspecific genetic variation of African fauna has been significantly affected by pronounced climatic fluctuations in Plio-Pleistocene, but, with the exception of large mammals, very limited empirical data on diversity of natural populations are available for savanna-dwelling animals. Nothobranchius furzeri is an annual fish from south-eastern Africa, inhabiting discrete temporary savannah pools outside main river alluvia. Their dispersal is limited and population processes affecting its genetic structure are likely a combination of those affecting terrestrial and aquatic taxa. N. furzeri is a model taxon in ageing research and several populations of known geographical origin are used in laboratory studies. Here, we analysed the genetic structure, diversity, historical demography and temporal patterns of divergence in natural populations of N. furzeri across its entire distribution range. Results: Genetic structure and historical demography of N. furzeri were analysed using a combination of mitochondrial (partial cytochrome b sequences, 687 bp) and nuclear (13 microsatellites) markers in 693 fish from 36 populations. Genetic markers consistently demonstrated strong population structuring and suggested two main genetic groups associated with river basins. The split was dated to the Pliocene (>2 Mya). The northern group inhabits savannah pools across the basin of the intermittent river Chefu in south-western Mozambique and eastern Zimbabwe.