Organization of the Class I Region of the Bovine Major
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Evidence for Differential Alternative Splicing in Blood of Young Boys With
Stamova et al. Molecular Autism 2013, 4:30 http://www.molecularautism.com/content/4/1/30 RESEARCH Open Access Evidence for differential alternative splicing in blood of young boys with autism spectrum disorders Boryana S Stamova1,2,5*, Yingfang Tian1,2,4, Christine W Nordahl1,3, Mark D Shen1,3, Sally Rogers1,3, David G Amaral1,3 and Frank R Sharp1,2 Abstract Background: Since RNA expression differences have been reported in autism spectrum disorder (ASD) for blood and brain, and differential alternative splicing (DAS) has been reported in ASD brains, we determined if there was DAS in blood mRNA of ASD subjects compared to typically developing (TD) controls, as well as in ASD subgroups related to cerebral volume. Methods: RNA from blood was processed on whole genome exon arrays for 2-4–year-old ASD and TD boys. An ANCOVA with age and batch as covariates was used to predict DAS for ALL ASD (n=30), ASD with normal total cerebral volumes (NTCV), and ASD with large total cerebral volumes (LTCV) compared to TD controls (n=20). Results: A total of 53 genes were predicted to have DAS for ALL ASD versus TD, 169 genes for ASD_NTCV versus TD, 1 gene for ASD_LTCV versus TD, and 27 genes for ASD_LTCV versus ASD_NTCV. These differences were significant at P <0.05 after false discovery rate corrections for multiple comparisons (FDR <5% false positives). A number of the genes predicted to have DAS in ASD are known to regulate DAS (SFPQ, SRPK1, SRSF11, SRSF2IP, FUS, LSM14A). In addition, a number of genes with predicted DAS are involved in pathways implicated in previous ASD studies, such as ROS monocyte/macrophage, Natural Killer Cell, mTOR, and NGF signaling. -

A Comprehensive Mapping of the Structure and Gene Organisation in the Sheep MHC Class I Region N
Siva Subramaniam et al. BMC Genomics (2015) 16:810 DOI 10.1186/s12864-015-1992-4 RESEARCH ARTICLE Open Access A comprehensive mapping of the structure and gene organisation in the sheep MHC class I region N. Siva Subramaniam1, EF Morgan1, JD Wetherall1, MJ Stear2,3* and DM Groth1 Abstract Background: The major histocompatibility complex (MHC) is a chromosomal region that regulates immune responsiveness in vertebrates. This region is one of the most important for disease resistance because it has been associated with resistance or susceptibility to a wide variety of diseases and because the MHC often accounts for more of the variance than other loci. Selective breeding for disease resistance is becoming increasingly common in livestock industries, and it is important to determine how this will influence MHC polymorphism and resistance to diseases that are not targeted for selection. However, in sheep the order and sequence of the protein coding genes is controversial. Yet this information is needed to determine precisely how the MHC influences resistance and susceptibility to disease. Methods: CHORI bacterial artificial chromosomes (BACs) known to contain sequences from the sheep MHC class I region were sub-cloned, and the clones partially sequenced. The resulting sequences were analysed and re-assembled to identify gene content and organisation within each BAC. The low resolution MHC class I physical map was then compared to the cattle reference genome, the Chinese Merino sheep MHC map published by Gao, et al. (2010) and the recently available sheep reference genome. Results: Immune related class I genes are clustered into 3 blocks; beta, kappa and a novel block not previously identified in other organisms. -

Integrating Single-Step GWAS and Bipartite Networks Reconstruction Provides Novel Insights Into Yearling Weight and Carcass Traits in Hanwoo Beef Cattle
animals Article Integrating Single-Step GWAS and Bipartite Networks Reconstruction Provides Novel Insights into Yearling Weight and Carcass Traits in Hanwoo Beef Cattle Masoumeh Naserkheil 1 , Abolfazl Bahrami 1 , Deukhwan Lee 2,* and Hossein Mehrban 3 1 Department of Animal Science, University College of Agriculture and Natural Resources, University of Tehran, Karaj 77871-31587, Iran; [email protected] (M.N.); [email protected] (A.B.) 2 Department of Animal Life and Environment Sciences, Hankyong National University, Jungang-ro 327, Anseong-si, Gyeonggi-do 17579, Korea 3 Department of Animal Science, Shahrekord University, Shahrekord 88186-34141, Iran; [email protected] * Correspondence: [email protected]; Tel.: +82-31-670-5091 Received: 25 August 2020; Accepted: 6 October 2020; Published: 9 October 2020 Simple Summary: Hanwoo is an indigenous cattle breed in Korea and popular for meat production owing to its rapid growth and high-quality meat. Its yearling weight and carcass traits (backfat thickness, carcass weight, eye muscle area, and marbling score) are economically important for the selection of young and proven bulls. In recent decades, the advent of high throughput genotyping technologies has made it possible to perform genome-wide association studies (GWAS) for the detection of genomic regions associated with traits of economic interest in different species. In this study, we conducted a weighted single-step genome-wide association study which combines all genotypes, phenotypes and pedigree data in one step (ssGBLUP). It allows for the use of all SNPs simultaneously along with all phenotypes from genotyped and ungenotyped animals. Our results revealed 33 relevant genomic regions related to the traits of interest. -

Aneuploidy: Using Genetic Instability to Preserve a Haploid Genome?
Health Science Campus FINAL APPROVAL OF DISSERTATION Doctor of Philosophy in Biomedical Science (Cancer Biology) Aneuploidy: Using genetic instability to preserve a haploid genome? Submitted by: Ramona Ramdath In partial fulfillment of the requirements for the degree of Doctor of Philosophy in Biomedical Science Examination Committee Signature/Date Major Advisor: David Allison, M.D., Ph.D. Academic James Trempe, Ph.D. Advisory Committee: David Giovanucci, Ph.D. Randall Ruch, Ph.D. Ronald Mellgren, Ph.D. Senior Associate Dean College of Graduate Studies Michael S. Bisesi, Ph.D. Date of Defense: April 10, 2009 Aneuploidy: Using genetic instability to preserve a haploid genome? Ramona Ramdath University of Toledo, Health Science Campus 2009 Dedication I dedicate this dissertation to my grandfather who died of lung cancer two years ago, but who always instilled in us the value and importance of education. And to my mom and sister, both of whom have been pillars of support and stimulating conversations. To my sister, Rehanna, especially- I hope this inspires you to achieve all that you want to in life, academically and otherwise. ii Acknowledgements As we go through these academic journeys, there are so many along the way that make an impact not only on our work, but on our lives as well, and I would like to say a heartfelt thank you to all of those people: My Committee members- Dr. James Trempe, Dr. David Giovanucchi, Dr. Ronald Mellgren and Dr. Randall Ruch for their guidance, suggestions, support and confidence in me. My major advisor- Dr. David Allison, for his constructive criticism and positive reinforcement. -

Pharmacodynamic Effects of Seliciclib, an Orally Administered
Cancer Therapy: Clinical Pharmacodynamic Effects of Seliciclib, an Orally Administered Cell Cycle Modulator, in Undifferentiated Nasopharyngeal Cancer Wen-Son Hsieh,4 Ross Soo,1Bee-Keow Peh,2 Thomas Loh,4 Difeng Dong,3 Donny Soh,5 Lim-SoonWong,3,5 Simon Green,6 Judy Chiao,6 Chun-Ying Cui,1Yo k e -F o n g L a i , 4 Soo-Chin Lee,1Benjamin Mow,1 Richie Soong,2 Manuel Salto-Tellez,2 and Boon-Cher Goh1, 2 Abstract Purpose: Cell cycle dysregulation resulting in expression of antiapoptotic genes and uncontrolled proliferation is a feature of undifferentiated nasopharyngeal carcinoma. The pharmacodynamic effects of seliciclib, a cyclin-dependent kinase (CDK) inhibitor, were studied in patients with nasopharyngeal carcinoma. Experimental Design: Patients with treatment-naI«ve locally advanced nasopharyngeal carci- noma received seliciclib at 800 mg or 400 mg twice daily on days1 to 3 and 8 to12.Paired tumor samples obtained at baseline and on day 13 were assessed by light microscopy, immunohisto- chemistry, and transcriptionalprofiling using real-time PCR low-density array consisting of apanel of 380 genes related to cell cycle inhibition, apoptosis, signal transduction, and cell proliferation. Results: At 800 mg bd, one patient experienced grade 3 liver toxicity and another had grade 2 vomiting; no significant toxicities were experienced in 13 patients treated at 400 mg bd. Seven of fourteen evaluable patients had clinical evidence of tumor reduction. Some of these responses were associated with increased tumor apoptosis, necrosis, and decreases in plasma EBV DNA posttreatment. Reduced protein expression of Mcl-1, cyclin D1, phosphorylated retinoblastoma protein pRB (T821), and significant transcriptional down-regulation of genes related to cellular proliferation and survival were shown in some patients posttreatment, indicative of cell cycle modulation by seliciclib, more specifically inhibition of cdk2/cyclin E, cdk7/cyclin H, and cdk9/cyclinT. -

A Comprehensive Transcriptome Analysis of Skeletal Muscles in Two Polish Pig Breeds Differing in Fat and Meat Quality Traits
Genetics and Molecular Biology, 41, 1, 125-136 (2018) Copyright © 2018, Sociedade Brasileira de Genética. Printed in Brazil DOI: http://dx.doi.org/10.1590/1678-4685-GMB-2016-0101 Research Article A comprehensive transcriptome analysis of skeletal muscles in two Polish pig breeds differing in fat and meat quality traits Katarzyna Piórkowska1 , Kacper òukowski3, Katarzyna Ropka-Molik1, Miros»aw Tyra2 and Artur Gurgul1 1Department of Animal Molecular Biology, National Research Institute of Animal Production, Balice, Poland. 2Department of Pig Breeding, National Research Institute of Animal Production, Balice, Poland. 3Department of Cattle Breeding, National Research Institute of Animal Production, Balice, Poland. Abstract Pork is the most popular meat in the world. Unfortunately, the selection pressure focused on high meat content led to a reduction in pork quality. The present study used RNA-seq technology to identify metabolic process genes related to pork quality traits and fat deposition. Differentially expressed genes (DEGs) were identified between pigs of Pulawska and Polish Landrace breeds for two the most important muscles (semimembranosus and longissimus dorsi). A total of 71 significant DEGs were reported: 15 for longissimus dorsi and 56 for semimembranosus muscles. The genes overexpressed in Pulawska pigs were involved in lipid metabolism (APOD, LXRA, LIPE, AP2B1, ENSSSCG00000028753 and OAS2) and proteolysis (CST6, CTSD, ISG15 and UCHL1). In Polish Landrace pigs, genes playing a role in biological adhesion (KIT, VCAN, HES1, SFRP2, CDH11, SSX2IP and PCDH17), actin cytoskeletal organisation (FRMD6, LIMK1, KIF23 and CNN1) and calcium ion binding (PVALB, CIB2, PCDH17, VCAN and CDH11) were transcriptionally more active. The present study allows for better understanding of the physiological processes associated with lipid metabolism and muscle fiber organization. -

AB-PN532 PPP1R11-Py64 Antibody Phosphosite-Specific Polyclonal Antibody for Monitoring the Phosphorylation of Human PPP1R11 (HCG V)
AB-PN532 PPP1R11-pY64 Antibody Phosphosite-specific polyclonal antibody for monitoring the phosphorylation of human PPP1R11 (HCG V) Address: 8755 Ash Street, Suite 1 Vancouver, British Columbia, Email: [email protected] Canada V6P 6T3 Phone: 604-323-2547 Target Protein Name Long: Protein phosphatase 1 regulatory subunit 11 HCG V; HCG-V; HCGV; Hemochromatosis candidate gene V protein; inhibitor-3; IPP3; MGC125741; MGC125742; MGC125743; PP1RB; PPP1R11; Protein phosphatase 1 regulatory subunit 11; protein phosphatase 1, regulatory Alias: (inhibitor) subunit 11; protein phosphatase 1, regulatory (inhibitor) subunit 11, isoform 1; Protein phosphatase inhibitor 3; t-complex-associated-testis- expressed 5; TCTE5; TCTEX5 UniProt ID: O60927 Sequence Predicted Mass (KDa): 13953 126 AA; O60927) Observed SDS-PAGE Mass (KDa): 18-23 Immunogen Antibody Immunogen Source: Human PPP1R11 (HCG V) sequence peptide Cat. No.: PE-04ADI99 Antibody Immunogen Sequence: CCI(pY)EKP(bA)C (bA) = beta-alanine Corresponds to amino acid residues C61 to P67; In the PPI_Ypi1 domain. This is Location in Target: the major in vivo phosphorylation site in PPP1R11. Peptide Type: For phosphosite-specific recognition of target. Target Phosphosite: Tyr-64 Production Antibody Host Species: Rabbit Antibody Type: Polyclonal Antibody Ig Isotype Clone Lot: Immunoglobulin G The immunizing peptide was produced by solid phase synthesis on a multipep peptide synthesizer and purified by reverse-phase hplc chromatography. Purity was assessed by analytical hplc and the amino acid sequence confirmed by mass spectrometry analysis. This peptide was coupled to KLH prior to immunization into rabbits. New Zealand White rabbits were subcutaneously Production Method: injected with KLH-coupled immunizing peptide every 4 weeks for 4 months. -
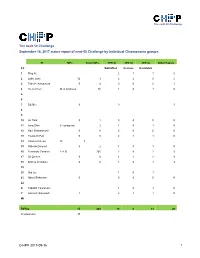
Next-MP50 Status Report
The neXt-50 Challenge The neXt-50 Challenge September 16, 2017 status report of next-50 Challenge by individual Chromosome groups. PI MPs Silver MPs JPR SI JPR SI JPR SI Other Papers Ch Submitted In press In revision 1 Ping Xu 2 1 1 0 2 Lydie Lane 12 3 2 2 0 2 3 Takeshi Kawamura 0 4 0 0 0 1 4 Yu-Ju Chen 26 in progress 86 1 0 1 0 5 6 7 Ed Nice 0 0 1 8 9 10 Jin Park 0 1 0 0 0 0 11 Jong Shin 2 in progress 2 1 0 1 0 12 Ravi Sirdeshmukh 0 0 0 0 0 0 13 Young-Ki Paik 0 5 2 1 1 0 14 CharLes Pineau 12 3 15 GiLberto Domont 0 2 1 0 1 0 16 Fernando CorraLes 2 (+ 9) 165 2 0 2 5 17 GiL Omenn 0 0 3 1 2 8 18 Andrey Archakov 0 0 1 0 1 3 19 20 Siqi Liu 1 0 1 21 Albert Sickmann 0 0 0 0 0 22 X Tadashi Yamamoto 1 0 1 0 Y Hosseini SaLekdeh 1 2 1 1 0 Mt TOTAL 15 268 19 6 13 20 (in progress) 37 C-HPP 2017-09-16 1 The neXt-50 Challenge Chromosome 1 (Ping Xu) PIC Leaders: Ping Xu, Fuchu He Contributing labs: Ping Xu, Beijing Proteome Research Center Fuchu He, Beijing Proteome Research Center Dong Yang, Beijing Proteome Research Center Wantao Ying, Beijing Proteome Research Center Pengyuan Yang, Fudan University Siqi Liu, Beijing Genome Institute Qinyu He, Jinan University Major lab members or partners contributing to the neXt50: Yao Zhang (Beijing Proteome Research Center), Yihao Wang (Beijing Proteome Research Center), Cuitong He (Beijing Proteome Research Center), Wei Wei (Beijing Proteome Research Center), Yanchang Li (Beijing Proteome Research Center), Feng Xu (Beijing Proteome Research Center), Xuehui Peng (Beijing Proteome Research Center). -

Transcriptional Recapitulation and Subversion Of
Open Access Research2007KaiseretVolume al. 8, Issue 7, Article R131 Transcriptional recapitulation and subversion of embryonic colon comment development by mouse colon tumor models and human colon cancer Sergio Kaiser¤*, Young-Kyu Park¤†, Jeffrey L Franklin†, Richard B Halberg‡, Ming Yu§, Walter J Jessen*, Johannes Freudenberg*, Xiaodi Chen‡, Kevin Haigis¶, Anil G Jegga*, Sue Kong*, Bhuvaneswari Sakthivel*, Huan Xu*, Timothy Reichling¥, Mohammad Azhar#, Gregory P Boivin**, reviews Reade B Roberts§, Anika C Bissahoyo§, Fausto Gonzales††, Greg C Bloom††, Steven Eschrich††, Scott L Carter‡‡, Jeremy E Aronow*, John Kleimeyer*, Michael Kleimeyer*, Vivek Ramaswamy*, Stephen H Settle†, Braden Boone†, Shawn Levy†, Jonathan M Graff§§, Thomas Doetschman#, Joanna Groden¥, William F Dove‡, David W Threadgill§, Timothy J Yeatman††, reports Robert J Coffey Jr† and Bruce J Aronow* Addresses: *Biomedical Informatics, Cincinnati Children's Hospital Medical Center, Cincinnati, OH 45229, USA. †Departments of Medicine, and Cell and Developmental Biology, Vanderbilt University and Department of Veterans Affairs Medical Center, Nashville, TN 37232, USA. ‡McArdle Laboratory for Cancer Research, University of Wisconsin, Madison, WI 53706, USA. §Department of Genetics and Lineberger Cancer Center, University of North Carolina, Chapel Hill, NC 27599, USA. ¶Molecular Pathology Unit and Center for Cancer Research, Massachusetts deposited research General Hospital, Charlestown, MA 02129, USA. ¥Division of Human Cancer Genetics, The Ohio State University College of Medicine, Columbus, Ohio 43210-2207, USA. #Institute for Collaborative BioResearch, University of Arizona, Tucson, AZ 85721-0036, USA. **University of Cincinnati, Department of Pathology and Laboratory Medicine, Cincinnati, OH 45267, USA. ††H Lee Moffitt Cancer Center and Research Institute, Tampa, FL 33612, USA. ‡‡Children's Hospital Informatics Program at the Harvard-MIT Division of Health Sciences and Technology (CHIP@HST), Harvard Medical School, Boston, Massachusetts 02115, USA. -

PP1RA Polyclonal Antibody Product Information
PP1RA Polyclonal Antibody Cat #: ABP59975 Size: 30μl /100μl /200μl Product Information Product Name: PP1RA Polyclonal Antibody Applications: WB, ELISA Isotype: Rabbit IgG Reactivity: Human, Mouse, Rat Catalog Number: ABP59975 Lot Number: Refer to product label Formulation: Liquid Concentration: 1 mg/ml Storage: Store at -20°C. Avoid repeated Note: Contain sodium azide. freeze / thaw cycles. Background: PPP1R10 (Protein Phosphatase 1 Regulatory Subunit 10) is a Protein Coding gene. Diseases associated with PPP1R10 include Malignant Melanoma, Somatic. Among its related pathways are Beta-Adrenergic Signaling and Activation of cAMP-Dependent PKA. PPP1R10 encodes a protein phosphatase 1 binding protein. The encoded protein plays a role in many cellular processes including cell cycle progression, DNA repair and apoptosis by regulating the activity of protein phosphatase 1. PPP1R10 lies within the major histocompatibility complex class I region on chromosome 6 and alternatively spliced transcript variants have been observed for PPP1R10. Application Notes: Optimal working dilutions should be determined experimentally by the investigator. Suggested starting dilutions are as follows: WB (1:500-1:2000), ELISA (1:5000-1:20000). Storage Buffer: PBS, pH 7.4, containing 0.02% Sodium Azide as preservative and 50% Glycerol. Storage Instructions: Stable for one year at -20°C from date of shipment. For maximum recovery of product, centrifuge the original vial after thawing and prior to removing the cap. Aliquot to avoid repeated freezing and thawing. Note: The product listed herein is for research use only and is not intended for use in human or clinical diagnosis. Suggested applications of our products are not recommendations to use our products in violation of any patent or as a license. -

Multiomic Analysis of the UV-Induced DNA Damage Response
Multiomic Analysis of the UV-Induced DNA Damage Response The MIT Faculty has made this article openly available. Please share how this access benefits you. Your story matters. Citation Boeing, Stefan, Laura Williamson, Vesela Encheva, Ilaria Gori, Rebecca E. Saunders, Rachael Instrell, Ozan Aygün, et al. “Multiomic Analysis of the UV-Induced DNA Damage Response.” Cell Reports 15, no. 7 (May 2016): 1597–1610. As Published http://dx.doi.org/10.1016/j.celrep.2016.04.047 Publisher Elsevier Version Final published version Citable link http://hdl.handle.net/1721.1/105271 Terms of Use Creative Commons Attribution 4.0 International License Detailed Terms http://creativecommons.org/licenses/by/4.0/ Resource Multiomic Analysis of the UV-Induced DNA Damage Response Graphical Abstract Authors Stefan Boeing, Laura Williamson, Vesela Encheva, ..., Michael Howell, Ambrosius P. Snijders, Jesper Q. Svejstrup Correspondence [email protected] In Brief Boeing et al. investigate the UV-induced DNA damage response by combining a range of proteomic and genomic screens. A function in this response for the melanoma driver STK19 as well as a number of other factors are uncovered. Highlights d A multiomic screening approach examines the UV-induced DNA damage response d Multiple factors are connected to the transcription-related DNA damage response d Melanoma gene STK19 is required for a normal DNA damage response Boeing et al., 2016, Cell Reports 15, 1597–1610 May 17, 2016 ª 2016 The Author(s) http://dx.doi.org/10.1016/j.celrep.2016.04.047 Cell Reports Resource Multiomic Analysis of the UV-Induced DNA Damage Response Stefan Boeing,1,5 Laura Williamson,1 Vesela Encheva,2 Ilaria Gori,3 Rebecca E. -

Lecture 15. Chromhmm & ENCODE
Lecture 15. ChromHMM & ENCODE Michael Schatz March 23, 2020 JHU 601.749: Applied Comparative Genomics Assignment 5: Due Mon Mar 23 Project Proposal: Due Mon Mar 23 *-seq in 4 short vignettes RNA-seq Methyl-seq ChIP-seq Hi-C Human Evolution ~5 Mya ~75 Mya ~100 Mya ~160 and 210 Mya As expected, the majority of platypus genes (82%; 15,312 out of 18,596) have orthologues in these five other amniotes (Supplementary Table 5). The remaining 'orphan' genes are expected to primarily reflect rapidly evolving genes, for which no other homologues are discernible, erroneous predictions, and true lineage-specific genes that have been lost in each of the other five species under consideration. Genome analysis of the platypus reveals unique signatures of evolution (2008) Nature. 453, 175-183 doi:10.1038/nature06936 Methyl-seq Finding the fifth base: Genome-wide sequencing of cytosine methylation Lister and Ecker (2009) Genome Research. 19: 959-966 Bisulfite Conversion Treating DNA with sodium bisulfite will convert unmethylated C to T • 5-MethyC will be protected and not change, so can look for differences when mapping • Requires great care when analyzing reads, since the complementary strand will also be converted (G to A) • Typically analyzed by mapping to a “reduced alphabet” where we assume all Cs are converted to Ts once on the forward strand and once on the reverse Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications Krueger and Andrews (2010) Bioinformatics. 27 (11): 1571-1572. ChIP-seq Genome-wide mapping of in vivo protein-DNA interactions. Johnson et al (2007) Science.