Identification of Qtls Affecting Scopolin and Scopoletin Biosynthesis in Arabidopsis Thaliana
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Scopoletin 8-Hydroxylase: a Novel Enzyme Involved in Coumarin
bioRxiv preprint doi: https://doi.org/10.1101/197806; this version posted October 4, 2017. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 1 Scopoletin 8-hydroxylase: a novel enzyme involved in coumarin biosynthesis and iron- 2 deficiency responses in Arabidopsis 3 4 Running title: At3g12900 encodes a scopoletin 8-hydroxylase 5 6 Joanna Siwinska1, Kinga Wcisla1, Alexandre Olry2,3, Jeremy Grosjean2,3, Alain Hehn2,3, 7 Frederic Bourgaud2,3, Andrew A. Meharg4, Manus Carey4, Ewa Lojkowska1, Anna 8 Ihnatowicz1,* 9 10 1Intercollegiate Faculty of Biotechnology of University of Gdansk and Medical University of 11 Gdansk, Abrahama 58, 80-307 Gdansk, Poland 12 2Université de Lorraine, UMR 1121 Laboratoire Agronomie et Environnement Nancy- 13 Colmar, 2 avenue de la forêt de Haye 54518 Vandœuvre-lès-Nancy, France; 3INRA, UMR 14 1121 Laboratoire Agronomie et Environnement Nancy-Colmar, 2 avenue de la forêt de Haye 15 54518 Vandœuvre-lès-Nancy, France; 16 4Institute for Global Food Security, Queen’s University Belfast, David Keir Building, Malone 17 Road, Belfast, UK; 18 19 [email protected] 20 [email protected] 21 [email protected] 22 [email protected] 23 [email protected] 24 [email protected] 25 [email protected] 26 [email protected] 27 [email protected] 28 *Correspondence: [email protected], +48 58 523 63 30 29 30 The date of submission: 02.10.2017 31 The number of figures: 9 (Fig. -

Biosynthesis of Daphnetin in Daphne Mezereum L.*
Biosynthesis of Daphnetin in Daphne mezereum L.* Stewart A. Brown Department of Chemistry, Trent University, Peterborough, Ont., Canada K9J 7B8 Z. Naturforsch. 41c, 247—252 (1986); received November 4, 1985 Biosynthesis, Coumarins, Daphne mezereum, Daphnetin Shoots of Daphne mezereum synthesized daphnetin (7,8-dihydroxycoumarin) more efficiently from [2-l4C]umbelliferone (7-hydroxycoumarin) than from [2-l4C]p-coumaric acid, and [2-14C]caf- feic acid was more poorly utilized still. These findings do not support the idea of derivation of daphnetin via hydroxylation of the caffeic acid ring at the 2 position, followed by lactone ring formation; instead they are consistent with the concept of daphnetin formation by an additional hydroxylation of umbelliferone at C-8. Umbelliferone was recovered with little l4C dilution from emulsin-hydrolysed extracts of shoots fed labelled umbelliferone, and TLC of extracts from un treated shoots revealed two substances yielding umbelliferone on hydrolysis. Evidence is pre sented from TLC and HPLC analysis that one of these is skimmin (7-O-ß-D-glucosylumbel- liferone), not previously reported from Daphne. The tracer experiments further support the theory that umbelliferone is the general precursor of coumarins bearing two or more hydroxyl functions on the aromatic ring. Introduction (Thymelaeaceae), a hardy perennial shrub, where it Approximately 70 coumarins with the 7,8 oxygen occurs together with its 7-ß-D-glucoside, daphnin ation pattern' have been reported to be naturally (6b) and daphnetin glucoside (6c), the 8-O-ß-D- occurring [1], over half being furanocoumarins de g lu c o s y la te d is o m e r [1]. -

A Detailed Review on Morphotaxonomy And
Acta Scientific Microbiology (ISSN: 2581-3226) Review Article Volume 1 Issue 6 June 2018 Skimmia anquetilia N.P. Taylor and Airy Shaw A Detailed Review on Morphotaxonomy and Chemoprofiling of Saduf Nissar1, Neelofar Majid1, Aabid M Rather1*, Irshad A Nawchoo1 and GG Mohi-Ud-Din2 1Plant Reproductive Biology, Genetic Diversity and Phytochemistry Research Laboratory, Department of Botany, University of Kashmir, Srinagar, India 2Department of Botany, Government Degree College for Women, Sopore, Baramullah, India *Corresponding Author: Aabid M Rather, Plant Reproductive Biology, Genetic Diversity and Phytochemistry Research Laboratory, DepartmentReceived: April of Botany, 20, 2018; University Published: of Kashmir, May 28, Srinagar, 2018 India. Abstract - In recent times, medicinal plants have attracted huge attention due to their diverse range of biological and therapeutic proper Skimmia ties. Evidences have been accumulated since ages to demonstrate promising potential of medicinal plants used in various traditional, anquetilia complementary, and alternative systems with the ever-increasing interest of today’s population towards natural products, Rutaceae N.P. Taylor and Airy Shaw emerged out to be one of the most eye-catching plant bearing multiple medicinal properties. It is a perennial aromatic evergreen shrub belonging to family . Pharmacological studies have demonstrated significant action - of different extracts as antimicrobial, anti-inflammatory agents, among others, supporting some of its popular uses. An attempt has S. anquetilia been made in this review article to provide an up-to-date overview of the morphological parameters, taxonomic features, distribu tion pattern, traditional uses, as well as the phytochemistry and biological activities of . The present review provides insights for future research aiming for both ethnopharmacological validation of its popular use and its exploration as a new source ofKeywords herbal drugs: Skimmia and/or anquetilia; bioactive Rutaceaenatural products. -
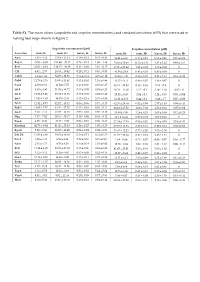
(Scopoletin and Scopolin Concentrations) and Standard Deviations (±SD) That Were Used in Making Heat Maps Shown in Figure 2
Table S1. The mean values (scopoletin and scopolin concentrations) and standard deviations (±SD) that were used in making heat maps shown in Figure 2. Scopoletin concentration [µM] Scopolin concentration [µM] Accession roots, H- roots, H+ leaves, H- leaves, H+ roots, H- roots, H+ leaves, H- leaves, H+ An-1 4.91 ± 4.12 15.19 ± 11.34 0.10 ± 0.12 0.45 ± 0.24 14.81 ± 4.05 0.13 ± 0.12 0.68 ± 0.41 0.05 ± 0.08 Bay-0 41.93 ± 6.58 151.90 ± 30.12 0.29 ± 0.18 1.99 ± 1.46 38.95 ± 10.69 07.15 ± 6.36 3.81 ± 1.22 0.88 ± 1.53 Br-0 35.01 ± 9.48 139.27 ± 49.38 0.31 ± 0.09 2.61 ± 0.27 67.26 ± 23.82 1.22 ± 0.08 1.65 ± 0.08 0 C24 4.92 ± 2.67 16.63 ± 10.42 0.20 ± 0.11 0.83 ± 0.55 41.36 ± 24.8 0.43 ± 0.19 0.85 ± 0.59 0 Can-0 3.14 ± 1.89 64.03 ± 84.91 0.14 ± 0.14 0.37 ± 0.12 34.63 ± 4.85 0.56 ± 0.05 2.23 ± 1.24 0.19 ± 0.33 Col-0 11.79 ± 3.16 29.60 ± 11.41 0.31 ± 0.41 1.16 ± 0.99 11.47 ± 3.41 0.46 ± 0.07 1.48 ± 0.67 0 Cvi-1 4.58 ± 0.62 11.54 ± 7.57 0.19 ± 0.10 0.61 ± 0.27 35.41 ± 32.83 0.35 ± 0.16 2.15 ± 0.8 0 Eil-0 8.03 ± 4.90 21.55 ± 14.72 0.15 ± 0.03 0.53 ± 0.23 28.24 ± 11.10 1.12 ± 0.2 2.19 ± 1.16 0.02 ± 0 Eri-0 12.86 ± 3.96 23.84 ± 13.33 0.13 ± 0.09 0.68 ± 0.48 20.55 ± 8.45 2.04 ± 0.3 2.24 ± 0.93 0.03 ± 0.04 Est-1 14.85 ± 1.63 36.79 ± 2.44 0.42 ± 0.19 2.01 ± 0.86 32.22 ±12.66 0.44 ± 0.2 6.48 ± 3.7 0.07 ± 0.09 Fei-0 12.82 ± 8.73 52.02 ± 15.32 0.09 ± 0.06 0.73 ± 0.37 62.36 ± 28.64 0.52 ± 0.08 2.77 ± 1.53 0.08 ± 0.13 Fuk-1 13.06 ± 7.87 44.36 ± 47.37 0.13 ± 0.01 0.62 ± 0.11 29.23 ± 27.52 8.09 ± 7.02 1.58 ± 0.62 0.05 ± 0.09 Ga-0 3.40 ± 1.11 19.67 ± 12.65 0.03 -

Phenylpropanoids
Phenylpropanoids The phenylpropanoids are a diverse family of organic compounds that are synthesized by plants from the amino acids phenylalanine and tyrosine. Their name is derived from the six-carbon, aromatic phenyl group and the three-carbon propene tail of cinnamic acid, which is synthesized from phenylalanine in the first step of phenylpropanoid biosynthesis. Phenylpropanoids are found throughout the plant kingdom, where they serve as essential components of a number of structural polymers, provide protection from ultraviolet light, defend against herbivores and pathogens, and mediate plant-pollinator interactions as floral pigments and scent compounds. Concentrations of phenylpropanoids within plants are also altered by changes in resource availability. www.MedChemExpress.com 1 Phenylpropanoids Inhibitors & Modulators (+)-Columbianetin (+)-Columbianetin acetate ((S)-Columbianetin) Cat. No.: HY-N0363 ((S)-Columbianetin acetate) Cat. No.: HY-N0363A (+)-Columbianetin is an isomer of Columbianetin. (S)-Columbianetin acetate is an isomer of Columbianetin is a phytoalexin associated with Columbianetin. Columbianetin is a phytoalexin celery (Apium graveolens) resistance to associated with celery (Apium graveolens) pathogens during storage. Columbianetin exhibits resistance to pathogens during storage. excellent anti-fungal and anti-inflammatory Columbianetin exhibits excellent anti-fungal and activity. anti-inflammatory activity. Purity: >98% Purity: >98% Clinical Data: No Development Reported Clinical Data: No Development Reported Size: 5 mg, 10 mg, 20 mg Size: 5 mg, 10 mg, 20 mg (+)-Guaiacin (+)-Peusedanol Cat. No.: HY-N2247A Cat. No.: HY-N6063 (+)-Guaiacin is a compound extracted of the bark (+)-Peusedanol is a coumarin isolated from of Machilus wangchiana Chun. (Lauraceae). Peucedanumjaponicum. (+)-Guaiacin shows potent in vitro activities against the release of β-glucuronidase in rat polymorphonuclear leukocytes (PMNs) induced by platelet-activating factor (PAF) . -

Scopoletin 8-Hydroxylase a Novel Enzyme Involved in Coumarin Biosynthesis and Iron-Deficiency Responses in Arabidopsis
Scopoletin 8-hydroxylase a novel enzyme involved in coumarin biosynthesis and iron-deficiency responses in Arabidopsis Siwinska, J., Siatkowska, K., Olry, A., Grosjean, J., Hehn, A., Bourgaud, F., Meharg, A. A., Carey, M., Lojkowska, E., & Ihnatowicz, A. (2018). Scopoletin 8-hydroxylase a novel enzyme involved in coumarin biosynthesis and iron-deficiency responses in Arabidopsis. Journal of experimental botany, 69(7), 1735-1748. https://doi.org/10.1093/jxb/ery005 Published in: Journal of experimental botany Document Version: Publisher's PDF, also known as Version of record Queen's University Belfast - Research Portal: Link to publication record in Queen's University Belfast Research Portal Publisher rights Copyright the authors 2018. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted reuse, distribution, and reproduction in any medium, provided the original work is properly cited. General rights Copyright for the publications made accessible via the Queen's University Belfast Research Portal is retained by the author(s) and / or other copyright owners and it is a condition of accessing these publications that users recognise and abide by the legal requirements associated with these rights. Take down policy The Research Portal is Queen's institutional repository that provides access to Queen's research output. Every effort has been made to ensure that content in the Research Portal does not infringe any person's rights, or applicable UK laws. If you discover content in the Research Portal that you believe breaches copyright or violates any law, please contact [email protected]. -

Identification of Qtls Affecting Scopolin and Scopoletin
Identification of QTLs affecting scopolin and scopoletin biosynthesis in Arabidopsis thaliana Joanna Siwinska, Leszek Kadzinski, Rafal Banasiuk, Anna Gwizdek-Wisniewska, Alexandre Olry, Bogdan Banecki, Ewa Lojkowska, Anna Ihnatowicz To cite this version: Joanna Siwinska, Leszek Kadzinski, Rafal Banasiuk, Anna Gwizdek-Wisniewska, Alexandre Olry, et al.. Identification of QTLs affecting scopolin and scopoletin biosynthesis in Arabidopsis thaliana. BMC Plant Biology, BioMed Central, 2014, 14, 10.1186/s12870-014-0280-9. hal-01477746 HAL Id: hal-01477746 https://hal.univ-lorraine.fr/hal-01477746 Submitted on 10 May 2017 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Siwinska et al. BMC Plant Biology 2014, 14:280 http://www.biomedcentral.com/1471-2229/14/280 RESEARCH ARTICLE Open Access Identification of QTLs affecting scopolin and scopoletin biosynthesis in Arabidopsis thaliana Joanna Siwinska1, Leszek Kadzinski1, Rafal Banasiuk1, Anna Gwizdek-Wisniewska1, Alexandre Olry2,3, Bogdan Banecki1, Ewa Lojkowska1 and Anna Ihnatowicz1* Abstract Background: Scopoletin and its glucoside scopolin are important secondary metabolites synthesized in plants as a defense mechanism against various environmental stresses. They belong to coumarins, a class of phytochemicals with significant biological activities that is widely used in medical application and cosmetics industry. -

Khellactone Derivatives and Other Phenolics of Phlojodicarpus Sibiricus (Apiaceae): HPLC-DAD-ESI-QQQ-MS/MS and HPLC-UV Profile
Article Khellactone Derivatives and Other Phenolics of Phlojodicarpus sibiricus (Apiaceae): HPLC‐DAD‐ESI‐QQQ‐MS/MS and HPLC‐UV Profile, and Antiobesity Potential of Dihydrosamidin Daniil N. Olennikov 1,*, Innokentii A. Fedorov 2, Nina I. Kashchenko 1, Nadezhda K. Chirikova 3 and Cecile Vennos 4 1 Institute of General and Experimental Biology, Siberian Division, Russian Academy of Science, 6 Sakh’yanovoy Street, Ulan‐Ude 670047, Russia; [email protected] 2 Institute for Biological Problems of Cryolithozone, Siberian Division, Russian Academy of Science, 41 Lenina Street, Yakutsk 677000, Russia; [email protected] 3 Department of Biochemistry and Biotechnology, North‐Eastern Federal University, 58 Belinsky Street, Yakutsk 677027, Russia; [email protected] 4 Regulatory and Medical Scientific Affairs, Padma AG, 1 Underfeldstrasse, Hinwil CH‐8340, Switzerland; [email protected] * Correspondence: [email protected]; Tel.: +7‐902‐160‐0627 Academic Editor: Alessandra Guerrini Received: 27 May 2019; Accepted: 18 June 2019; Published: 19 June 2019 Abstract: With obesity, the consumption of phenolic‐enriched food additives as a part of traditional nutrition avoids the negative implications of eating high‐calorie products. This study investigated the new herbal food additive, Phlojodicarpus sibiricus roots and herb, ubiquitously used in Siberia as a spice. Chromatographic techniques such as HPLC‐DAD‐ESI‐QQQ‐MS/MS and microcolumn HPLC‐UV were the basic instruments for component profiling and quantification, and antiobesity potential was investigated using a differentiated 3T3‐L1 adipocytes assay. We found that the roots and herb of P. sibiricus were high‐coumarin‐containing additives inhibiting triacylglycerol accumulation in 3T3‐L1 preadipocytes. Forty‐one phenolics were detected in P. -

Two Glucosides of Coumarin Derivatives in Sweet Potato Roots Infected by Ceratocystis Fimbriata by Takao MINAMIKAWA, TAKASHI
[Agr. Biol. Chem., Vol. 28, No. 4, p. 230•`233, 1964] Two Glucosides of Coumarin Derivatives in Sweet Potato Roots Infected by Ceratocystis Fimbriata By Takao MINAMIKAWA,TAKASHI AKAZAWA and IKUZO URITANI Laboratoryof BiochemistryNagoya University, Anjo, Aichi ReceivedJanuary 13, 1964 Two glucosides of coumarin derivatives were separated from sweet potato roots with black rot, by the combination of silica gel-coated thin layer chromatography and paper chromatography, and identified with skimmin and scopolin. The magnitude of synthesis of both bound coumarins was lower than that of the corresponding free coumarins, whereas they were detectable in neither cut nor fresh root tissues. sues7). During the course of this work, we INTRODUCTION also tried to find out whether or not the Several workers have reported the produc fungus infection would induce the produc tion of coumarins in plant tissues infected by tion of the bound forms of the respective pathogenic microorganisms1•`4) Post-infec coumarins in the root tissues. Hughes and tion synthesis of umbelliferone (7-hydroxy- Swain4) reported that about 25-fold increase coumarin) and scopoletin (6-methoxy-7-hy of scopolin (scopoletin-7-ƒÀ-glucoside) was in droxycoumarin) in sweet potato roots attack duced by the infection of Phytophthora in ed by the black rot fungus, Ceratocystis festans in potato tubers, whereas the chloro fimbriata, was reported by Uritani and Hoshi genic acid increase was only 2- to 5-fold in - ya5), and recently the third coumarin com the infected region. pound, esculetin (6,7-dihydroxycoumarin), This paper describes the chromatographic has been added to the list6). We have also identification of skimmin (umbelliferone-7-ƒÀ- carried out an analytical study on the synthe sis of the former two coumarins in the in glucoside) and scopolin in the sweet potato tissue with black rot. -

Coumarins and Coumarin-Related Compounds in Pharmacotherapy of Cancer
cancers Review Coumarins and Coumarin-Related Compounds in Pharmacotherapy of Cancer Esra Küpeli Akkol 1,* , Yasin Genç 2, Bü¸sraKarpuz 1, Eduardo Sobarzo-Sánchez 3,4 and Raffaele Capasso 5,* 1 Department of Pharmacognosy, Faculty of Pharmacy, Gazi University, Etiler 06330, Ankara, Turkey; [email protected] 2 Department of Pharmacognosy, Faculty of Pharmacy, Hacettepe University, Sıhhiye 06100, Ankara, Turkey; [email protected] 3 Instituto de Investigación e Innovación en Salud, Facultad de Ciencias de la Salud, Universidad Central de Chile, 8330507 Santiago, Chile; [email protected] 4 Department of Organic Chemistry, Faculty of Pharmacy, University of Santiago de Compostela, 15782 Santiago de Compostela, Spain 5 Department of Agricultural Sciences, University of Naples Federico II, 80055 Portici (Naples), Italy * Correspondence: [email protected] (E.K.A.); [email protected] (R.C.); Tel.: +90-312-2023185 (E.K.A); +39-081-678664 (R.C.) Received: 3 July 2020; Accepted: 17 July 2020; Published: 19 July 2020 Abstract: Cancer is one of the most common causes of disease-related deaths worldwide. Despite the discovery of many chemotherapeutic drugs that inhibit uncontrolled cell division processes for the treatment of various cancers, serious side effects of these drugs are a crucial disadvantage. In addition, multi-drug resistance is another important problem in anticancer treatment. Due to problems such as cytotoxicity and drug resistance, many investigations are being conducted to discover and develop effective anticancer drugs. In recent years, researchers have focused on the anticancer activity coumarins, due to their high biological activity and low toxicity. Coumarins are commonly used in the treatment of prostate cancer, renal cell carcinoma and leukemia, and they also have the ability to counteract the side effects caused by radiotherapy. -

Coumarins and Metabolic Syndrome: Brief Report RESEARCH ARTICLE
Mariana Alonso. et al. Medical Research Archives vol 5 Issue 11. November Issue. Medical Research Archives RESEARCH ARTICLE Coumarins and metabolic syndrome: Brief Report Authors Mariana Macias Alonso,1* Joaquín G. Marrero,1* Iván Córdova Guerrero,2 Claudia Pérez Sánchez,1 Soraya Osegueda Robles.1 Affiliations 1 Instituto Politécnico Nacional, UPIIG, Av. Mineral de Valenciana, No. 200, Col. Fracc. Industrial Puerto Interior, C.P. 36275 Silao de la Victoria, Guanajuato, México 2 Facultad de Ciencias Químicas e Ing. Universidad Autónoma de Baja California. Clz. Universidad 14418, Parque Industrial Internacional. Tijuana, B. C. CP 22390. México Correspondence authors: Mariana Macías Alonso; E-mail: [email protected] Joaquín González Marrero E-mail: [email protected] Abstract Metabolic syndrome is defined by the presentation of a wide array of interconnected physiological, biochemical, clinical and metabolic abnormalities that mainly increase the risk of type 2 diabetes mellitus and cardiovascular disease, which is commonly recognized as the primary clinical outcome. Pharmacological treatment for those whose risk factors are not adequately reduced with lifestyle changes remains challenging due to the polypharmacy, the risk of side effects and interactions especially in long-term treatments. The aim of this study was a systematic review of the literature published in previous years about the effects of coumarins against metabolic syndrome using in-vivo animal models. All studies included where pharmacological treatment was given to animals with obesity or diabetes mellitus. Studies included at least 90% of guidelines of the Gold Standard Publication Checklist (GSPC) for improved design, reporting and scientific quality of animal studies. Twenty studies reporting on the effects of different pharmacological treatments were included. -

WO 2018/046526 Al 15 March 2018 (15.03.2018) W !P O PCT
(12) INTERNATIONAL APPLICATION PUBLISHED UNDER THE PATENT COOPERATION TREATY (PCT) (19) World Intellectual Property Organization International Bureau (10) International Publication Number (43) International Publication Date WO 2018/046526 Al 15 March 2018 (15.03.2018) W !P O PCT (51) International Patent Classification: CI2N 15/82 (2006.01) (21) International Application Number: PCT/EP20 17/0723 17 (22) International Filing Date: 06 September 2017 (06.09.2017) (25) Filing Language: English (26) Publication Language: English (30) Priority Data: 16187335.1 06 September 2016 (06.09.2016) EP (71) Applicants: VIB VZW [BE/BE]; Rijvisschestraat 120, 9052 Gent (BE). UNIVERSITEIT GENT [BE/BE]; Sint- Pietersnieuwstraat 25, 9000 Gent (BE). (72) Inventors: BOERJAN, Wout; Zomerstraat 44B, 9270 Kalken (BE). SUNDIN, Lisa; Sint-Jansdreef 17, 9000 Gent (BE). VANHOLME, Ruben; Panhuisstraat 30, 9070 Destelbergen (BE). = (74) Common Representative: VIB VZW; Rijvisschestraat = 120, 9052 Gent (BE). (81) Designated States (unless otherwise indicated, for every kind of national protection available): AE, AG, AL, AM, = AO, AT, AU, AZ, BA, BB, BG, BH, BN, BR, BW, BY, BZ, = CA, CH, CL, CN, CO, CR, CU, CZ, DE, DJ, DK, DM, DO, ≡ DZ, EC, EE, EG, ES, FI, GB, GD, GE, GH, GM, GT, HN, = HR, HU, ID, IL, IN, IR, IS, JO, JP, KE, KG, KH, KN, KP, = KR, KW, KZ, LA, LC, LK, LR, LS, LU, LY, MA, MD, ME, — MG, MK, MN, MW, MX, MY, MZ, NA, NG, NI, NO, NZ, = OM, PA, PE, PG, PH, PL, PT, QA, RO, RS, RU, RW, SA, = SC, SD, SE, SG, SK, SL, SM, ST, SV, SY, TH, TJ, TM, TN, ≡ TR, TT, TZ, UA, UG, US, UZ, VC, VN, ZA, ZM, ZW.