Supporting Online Material (SOM)
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Molecular Genetic Analysis of Two Loci (Ity2 and Ity3) Involved in The
Copyright Ó 2007 by the Genetics Society of America DOI: 10.1534/genetics.107.075523 Molecular Genetic Analysis of Two Loci (Ity2 and Ity3) Involved in the Host Response to Infection With Salmonella Typhimurium Using Congenic Mice and Expression Profiling Vanessa Sancho-Shimizu,*,† Rabia Khan,*,† Serge Mostowy,† Line Larivie`re,† Rosalie Wilkinson,† Noe´mie Riendeau,† Marcel Behr† and Danielle Malo*,†,‡,1 *Department of Human Genetics, McGill University, Montreal, Quebec H3G 1A4, Canada and †Center for the Study of Host Resistance, McGill University Health Center, Montreal, Quebec H3G 1A4, Canada and ‡Department of Medicine, McGill University, Montreal, Quebec H3G 1A4, Canada Manuscript received May 4, 2007 Accepted for publication July 27, 2007 ABSTRACT Numerous genes have been identified to date that contribute to the host response to systemic Sal- monella Typhimurium infection in mice. We have previously identified two loci, Ity2 and Ity3, that control survival to Salmonella infection in the wild-derived inbred MOLF/Ei mouse using a (C57BL/6J 3 MOLF/ Ei)F2cross. We validated the existence of these two loci by creating congenic mice carrying each quan- titative trait locus (QTL) in isolation. Subcongenic mice generated for each locus allowed us to define the critical intervals underlying Ity2 and Ity3. Furthermore, expression profiling was carried out with the aim of identifying differentially expressed genes within the critical intervals as potential candidate genes. Genomewide expression arrays were used to interrogate expression differences in the Ity2 congenics, leading to the identification of a new candidate gene (Havcr2, hepatitis A virus cellular receptor 2). Interval-specific oligonucleotide arrays were created for Ity3, identifying one potential candidate gene (Chi3l1, chitinase 3-like 1) to be pursued further. -

The Rise and Fall of the Bovine Corpus Luteum
University of Nebraska Medical Center DigitalCommons@UNMC Theses & Dissertations Graduate Studies Spring 5-6-2017 The Rise and Fall of the Bovine Corpus Luteum Heather Talbott University of Nebraska Medical Center Follow this and additional works at: https://digitalcommons.unmc.edu/etd Part of the Biochemistry Commons, Molecular Biology Commons, and the Obstetrics and Gynecology Commons Recommended Citation Talbott, Heather, "The Rise and Fall of the Bovine Corpus Luteum" (2017). Theses & Dissertations. 207. https://digitalcommons.unmc.edu/etd/207 This Dissertation is brought to you for free and open access by the Graduate Studies at DigitalCommons@UNMC. It has been accepted for inclusion in Theses & Dissertations by an authorized administrator of DigitalCommons@UNMC. For more information, please contact [email protected]. THE RISE AND FALL OF THE BOVINE CORPUS LUTEUM by Heather Talbott A DISSERTATION Presented to the Faculty of the University of Nebraska Graduate College in Partial Fulfillment of the Requirements for the Degree of Doctor of Philosophy Biochemistry and Molecular Biology Graduate Program Under the Supervision of Professor John S. Davis University of Nebraska Medical Center Omaha, Nebraska May, 2017 Supervisory Committee: Carol A. Casey, Ph.D. Andrea S. Cupp, Ph.D. Parmender P. Mehta, Ph.D. Justin L. Mott, Ph.D. i ACKNOWLEDGEMENTS This dissertation was supported by the Agriculture and Food Research Initiative from the USDA National Institute of Food and Agriculture (NIFA) Pre-doctoral award; University of Nebraska Medical Center Graduate Student Assistantship; University of Nebraska Medical Center Exceptional Incoming Graduate Student Award; the VA Nebraska-Western Iowa Health Care System Department of Veterans Affairs; and The Olson Center for Women’s Health, Department of Obstetrics and Gynecology, Nebraska Medical Center. -

What Are Their Roles in Mitochondrial Protein Synthesis?
Characterisation of human mtRF1 and C12orf65: What are their roles in mitochondrial protein synthesis? Aleksandra Pajak M.Res Thesis submitted to Newcastle University in candidature for the degree of Doctor of Philosophy Newcastle University Faculty of Medical Sciences Institute for Ageing and Health Mitochondrial Research Group January 2013 Abstract Mitochondria have their own protein synthesis machinery that synthesises the oxidative phosphorylation components encoded by their mtDNA. This translation process consists of four main phases: initiation, elongation, termination and ribosome recycling. Termination and its control have been the least investigated. Recently, however, the termination factor, mtRF1a, has been characterised as sufficient to release all the nascent proteins from the mitoribosome. Furthermore, bioinformatics has identified three additional members of this mitochondrial release factor family namely, mtRF1, C12orf65 and ICT1. The latter is now known to be incorporated into the mitoribosome but its exact function remains unclear. My project has therefore focussed on characterising the remaining two factors; mtRF1 and C12orf65, and investigating their possible involvement in mitochondrial protein synthesis. It has been demonstrated that protein synthesis is not perfect and bacterial ribosomes not infrequently stall during translation. This can result from limiting amounts of charged tRNAs, stable secondary structures, or truncated/degraded transcripts. Ribosome stalling has been shown to cause growth arrest. In order to prevent that and maintain high efficiency of mitochondrial protein synthesis such stalled complexes need to be rapidly recycled. Bacteria have developed at least three distinct mechanisms by which ribosomes can be rescued. Contrastingly, despite the presence of truncated mRNAs in mitochondria, no such quality control mechanisms have been identified in these organelles. -

Table 2. Significant
Table 2. Significant (Q < 0.05 and |d | > 0.5) transcripts from the meta-analysis Gene Chr Mb Gene Name Affy ProbeSet cDNA_IDs d HAP/LAP d HAP/LAP d d IS Average d Ztest P values Q-value Symbol ID (study #5) 1 2 STS B2m 2 122 beta-2 microglobulin 1452428_a_at AI848245 1.75334941 4 3.2 4 3.2316485 1.07398E-09 5.69E-08 Man2b1 8 84.4 mannosidase 2, alpha B1 1416340_a_at H4049B01 3.75722111 3.87309653 2.1 1.6 2.84852656 5.32443E-07 1.58E-05 1110032A03Rik 9 50.9 RIKEN cDNA 1110032A03 gene 1417211_a_at H4035E05 4 1.66015788 4 1.7 2.82772795 2.94266E-05 0.000527 NA 9 48.5 --- 1456111_at 3.43701477 1.85785922 4 2 2.8237185 9.97969E-08 3.48E-06 Scn4b 9 45.3 Sodium channel, type IV, beta 1434008_at AI844796 3.79536664 1.63774235 3.3 2.3 2.75319499 1.48057E-08 6.21E-07 polypeptide Gadd45gip1 8 84.1 RIKEN cDNA 2310040G17 gene 1417619_at 4 3.38875643 1.4 2 2.69163229 8.84279E-06 0.0001904 BC056474 15 12.1 Mus musculus cDNA clone 1424117_at H3030A06 3.95752801 2.42838452 1.9 2.2 2.62132809 1.3344E-08 5.66E-07 MGC:67360 IMAGE:6823629, complete cds NA 4 153 guanine nucleotide binding protein, 1454696_at -3.46081884 -4 -1.3 -1.6 -2.6026947 8.58458E-05 0.0012617 beta 1 Gnb1 4 153 guanine nucleotide binding protein, 1417432_a_at H3094D02 -3.13334396 -4 -1.6 -1.7 -2.5946297 1.04542E-05 0.0002202 beta 1 Gadd45gip1 8 84.1 RAD23a homolog (S. -

Evaluation of Cancer-Derived Myocardial Impairments Using a Mouse Model
www.oncotarget.com Oncotarget, 2020, Vol. 11, (No. 41), pp: 3712-3722 Research Paper Evaluation of cancer-derived myocardial impairments using a mouse model Yoshihiro Miyagawa1, Shota Nukaga1,2, Takuya Mori1, Rina Fujiwara-Tani1, Kiyomu Fujii1, Shiori Mori1, Kei Goto1,3, Shingo Kishi1, Takamitsu Sasaki1, Chie Nakashima1, Hitoshi Ohmori1, Isao Kawahara1,2, Yi Luo4 and Hiroki Kuniyasu1 1Department of Molecular Pathology, Nara Medical University, Kashihara, Nara 634-8521, Japan 2Division of Rehabilitation, Hanna Central Hospital, Ikoma, Nara 630-0243, Japan 3Division of Rehabilitation, Hoshida Minami Hospital, Katano, Osaka 576-0022, Japan 4Key Laboratory of Neuroregeneration of Jiangsu and Ministry of Education, Co-Innovation Center of Neuroregeneration, Nantong University, Nantong, Jiangsu Province 226001, China Correspondence to: Yi Luo, email: [email protected] Hiroki Kuniyasu, email: [email protected] Keywords: cachexia; myocardium; atrophy; mitochondria; oxidative stress Received: June 26, 2020 Accepted: September 10, 2020 Published: October 13, 2020 Copyright: © 2020 Miyagawa et al. This is an open access article distributed under the terms of the Creative Commons Attribution License (CC BY 3.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. ABSTRACT Myocardial damage in cancer patients is emphasized as a cause of death; however, there are not many murine cachexia models to evaluate cancer-derived heart disorder. Using the mouse cachexia model that we established previously, we investigated myocardial damage in tumor-bearing mice. In cachexic mice, decreased heart weight and myocardial volume, and dilated left ventricular lumen, and atrophied cardiomyocytes were noted. The cardiomyocytes also showed accumulated 8-hydroxydeoxyguanosine, decreased leucine zipper and EF-hand- containing transmembrane protein-1, and increased microtubule-associated protein light chain3-II. -

Greg's Awesome Thesis
Analysis of alignment error and sitewise constraint in mammalian comparative genomics Gregory Jordan European Bioinformatics Institute University of Cambridge A dissertation submitted for the degree of Doctor of Philosophy November 30, 2011 To my parents, who kept us thinking and playing This dissertation is the result of my own work and includes nothing which is the out- come of work done in collaboration except where specifically indicated in the text and acknowledgements. This dissertation is not substantially the same as any I have submitted for a degree, diploma or other qualification at any other university, and no part has already been, or is currently being submitted for any degree, diploma or other qualification. This dissertation does not exceed the specified length limit of 60,000 words as defined by the Biology Degree Committee. November 30, 2011 Gregory Jordan ii Analysis of alignment error and sitewise constraint in mammalian comparative genomics Summary Gregory Jordan November 30, 2011 Darwin College Insight into the evolution of protein-coding genes can be gained from the use of phylogenetic codon models. Recently sequenced mammalian genomes and powerful analysis methods developed over the past decade provide the potential to globally measure the impact of natural selection on pro- tein sequences at a fine scale. The detection of positive selection in particular is of great interest, with relevance to the study of host-parasite conflicts, immune system evolution and adaptive dif- ferences between species. This thesis examines the performance of methods for detecting positive selection first with a series of simulation experiments, and then with two empirical studies in mammals and primates. -

LETM1 Gene Leucine Zipper and EF-Hand Containing Transmembrane Protein 1
LETM1 gene leucine zipper and EF-hand containing transmembrane protein 1 Normal Function The LETM1 gene provides instructions for making a protein whose function is not well understood. This protein is active in mitochondria, which are structures within cells that convert the energy from food into a form that cells can use. The LETM1 protein may be involved in the transport of charged calcium atoms (calcium ions) across membranes within mitochondria. Researchers suspect that the protein also plays a role in determining the shape and volume of mitochondria. Health Conditions Related to Genetic Changes Wolf-Hirschhorn syndrome The LETM1 gene is located in a region of chromosome 4 that is deleted in people with the typical features of Wolf-Hirschhorn syndrome. As a result of this deletion, affected individuals are missing one copy of the LETM1 gene in each cell. Studies suggest that a loss of this gene alters the structure of mitochondria; however, it is unclear how this abnormality is related to the signs and symptoms of Wolf-Hirschhorn syndrome. Specifically, a loss of the LETM1 gene has been associated with seizures or other abnormal electrical activity in the brain. Other Names for This Gene • LETM1_HUMAN • leucine zipper-EF-hand containing transmembrane protein 1 Additional Information & Resources Tests Listed in the Genetic Testing Registry • Tests of LETM1 (https://www.ncbi.nlm.nih.gov/gtr/all/tests/?term=3954[geneid]) Scientific Articles on PubMed Reprinted from MedlinePlus Genetics (https://medlineplus.gov/genetics/) 1 • PubMed (https://pubmed.ncbi.nlm.nih.gov/?term=%28%28LETM1%5BTIAB%5D%2 -

LETM1 Haploinsufficiency Causes Mitochondrial Defects in Cells From
© 2014. Published by The Company of Biologists Ltd | Disease Models & Mechanisms (2014) 7, 535-545 doi:10.1242/dmm.014464 RESEARCH ARTICLE LETM1 haploinsufficiency causes mitochondrial defects in cells from humans with Wolf-Hirschhorn syndrome: implications for dissecting the underlying pathomechanisms in this condition Lesley Hart1,2, Anita Rauch3, Antony M. Carr2, Joris R. Vermeesch4 and Mark O’Driscoll1,* ABSTRACT abnormalities, a characteristic facial dysmorphology, hypotonia, and Wolf-Hirschhorn syndrome (WHS) represents an archetypical epileptic seizures (Hirschhorn and Cooper, 1961; Hirschhorn et al., example of a contiguous gene deletion disorder – a condition 1965; Wolf et al., 1965). The spectrum and severity of these clinical comprising a complex set of developmental phenotypes with a features typically correlate with deletion size (Battaglia et al., 2008; multigenic origin. Epileptic seizures, intellectual disability, growth Maas et al., 2008; Van Buggenhout et al., 2004; Zollino et al., 2000). restriction, motor delay and hypotonia are major co-morbidities in WHS is generally regarded as a multigenic disorder, although two WHS. Haploinsufficiency of LETM1, which encodes a mitochondrial critical regions have been described: WHSCR1 and WHSCR2, for inner-membrane protein functioning in ion transport, has been WHS critical region 1 and 2, respectively (see Fig. 1). These critical proposed as an underlying pathomechanism, principally for seizures regions are based on the demarcation of the minimum region of but also for other core features of WHS, including growth and motor overlap in individuals exhibiting WHS-like phenotypes. WHSCR1 delay. Growing evidence derived from several model organisms incorporates part of the WHS candidate gene WHSC1 and the entire suggests that reduced LETM1 expression is associated with some WHSC2 gene (White et al., 1995; Wright et al., 1997). -

Stríbrná and Julius Lukes Hassan Hashimi, Lindsay Mcdonald, Eva
Bioenergetics: Trypanosome Letm1 Protein Is Essential for Mitochondrial Potassium Homeostasis Hassan Hashimi, Lindsay McDonald, Eva Stríbrná and Julius Lukes J. Biol. Chem. 2013, 288:26914-26925. doi: 10.1074/jbc.M113.495119 originally published online July 26, 2013 Access the most updated version of this article at doi: 10.1074/jbc.M113.495119 Find articles, minireviews, Reflections and Classics on similar topics on the JBC Affinity Sites. Alerts: • When this article is cited • When a correction for this article is posted Click here to choose from all of JBC's e-mail alerts Supplemental material: http://www.jbc.org/content/suppl/2013/07/26/M113.495119.DC1.html This article cites 60 references, 26 of which can be accessed free at http://www.jbc.org/content/288/37/26914.full.html#ref-list-1 Downloaded from http://www.jbc.org/ at Edinburgh University Library on September 13, 2013 THE JOURNAL OF BIOLOGICAL CHEMISTRY VOL. 288, NO. 37, pp. 26914–26925, September 13, 2013 © 2013 by The American Society for Biochemistry and Molecular Biology, Inc. Published in the U.S.A. Trypanosome Letm1 Protein Is Essential for Mitochondrial Potassium Homeostasis*□S Received for publication, June 19, 2013, and in revised form, July 23, 2013 Published, JBC Papers in Press, July 26, 2013, DOI 10.1074/jbc.M113.495119 Hassan Hashimi‡§1, Lindsay McDonald‡2, Eva Strˇíbrná‡, and Julius Lukesˇ‡§3 From the ‡Institute of Parasitology, Biology Centre, Czech Academy of Sciences and the §Faculty of Science, University of South Bohemia, 370 05 Cˇeské Budeˇjovice (Budweis), Czech Republic Background: Letm1 is a mitochondrial protein attributed disparate roles, including cation/proton antiport and translation. -

Association of Interleukin-10 Cluster Genes and Salmonella Response in the Chicken
Animal Science Publications Animal Science 1-1-2008 Association of Interleukin-10 Cluster Genes and Salmonella Response in the Chicken S. B. Ghebremicael Iowa State University J. Hasenstein Iowa State University Susan J. Lamont Iowa State University, [email protected] Follow this and additional works at: https://lib.dr.iastate.edu/ans_pubs Part of the Agriculture Commons, Genetics Commons, and the Poultry or Avian Science Commons The complete bibliographic information for this item can be found at https://lib.dr.iastate.edu/ ans_pubs/607. For information on how to cite this item, please visit http://lib.dr.iastate.edu/ howtocite.html. This Article is brought to you for free and open access by the Animal Science at Iowa State University Digital Repository. It has been accepted for inclusion in Animal Science Publications by an authorized administrator of Iowa State University Digital Repository. For more information, please contact [email protected]. Association of Interleukin-10 Cluster Genes and Salmonella Response in the Chicken Abstract Salmonella enteritidis lipopolysaccharide stimulates interleukin 10 (IL10) gene expression in chickens. Four genes in the IL10 cluster [polymeric immunoglobulin receptor (PIGR), interleukin 10 (IL10), map kinase-activated protein kinase 2 (MAPKAPK2), and ligatin (LGTN)] plus dual-specificity tyrosine-(Y)- phosphorylation regulated kinase1A (DYRK1A) were investigated using the F8 generation of 2 related advanced intercross lines (AIL). The AIL were generated by crossing outbred broilers with dams of 2 highly inbred lines (Leghorn and Fayoumi). Intercrossing continued within the 2 dam lines. The F8 chicks (n = 132) were intraesophageally inoculated at 1 d with S. enteritidis. At d 7 or 8, both spleen tissue and cecal contents were cultured to quantify S. -

Genome-Wide Investigation of Cellular Functions for Trna Nucleus
Genome-wide Investigation of Cellular Functions for tRNA Nucleus- Cytoplasm Trafficking in the Yeast Saccharomyces cerevisiae DISSERTATION Presented in Partial Fulfillment of the Requirements for the Degree Doctor of Philosophy in the Graduate School of The Ohio State University By Hui-Yi Chu Graduate Program in Molecular, Cellular and Developmental Biology The Ohio State University 2012 Dissertation Committee: Anita K. Hopper, Advisor Stephen Osmani Kurt Fredrick Jane Jackman Copyright by Hui-Yi Chu 2012 Abstract In eukaryotic cells tRNAs are transcribed in the nucleus and exported to the cytoplasm for their essential role in protein synthesis. This export event was thought to be unidirectional. Surprisingly, several lines of evidence showed that mature cytoplasmic tRNAs shuttle between nucleus and cytoplasm and their distribution is nutrient-dependent. This newly discovered tRNA retrograde process is conserved from yeast to vertebrates. Although how exactly the tRNA nuclear-cytoplasmic trafficking is regulated is still under investigation, previous studies identified several transporters involved in tRNA subcellular dynamics. At least three members of the β-importin family function in tRNA nuclear-cytoplasmic intracellular movement: (1) Los1 functions in both the tRNA primary export and re-export processes; (2) Mtr10, directly or indirectly, is responsible for the constitutive retrograde import of cytoplasmic tRNA to the nucleus; (3) Msn5 functions solely in the re-export process. In this thesis I focus on the physiological role(s) of the tRNA nuclear retrograde pathway. One possibility is that nuclear accumulation of cytoplasmic tRNA serves to modulate translation of particular transcripts. To test this hypothesis, I compared expression profiles from non-translating mRNAs and polyribosome-bound translating mRNAs collected from msn5Δ and mtr10Δ mutants and wild-type cells, in fed or acute amino acid starvation conditions. -
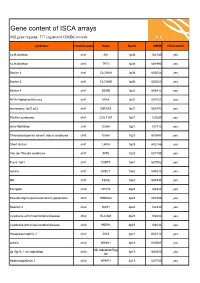
ISCA Disease List
Gene content of ISCA arrays 402 gene regions, 377 registered OMIM records syndrome chromosome Gene Band OMIM ISCA 8x60k 1p36 deletion chr1 SKI 1p36 164780 yes 1p36 deletion chr1 TP73 1p36 601990 yes Bartter 4 chr1 CLCNKA 1p36 602024 yes Bartter 3 chr1 CLCNKB 1p36 602023 yes Bartter 4 chr1 BSND 1p32 606412 yes NFIA Haploinsufficiency chr1 NFIA 1p31 600727 yes monosomy 1p31 p22 chr1 DIRAS3 1p31 605193 yes Stickler syndrome chr1 COL11A1 1p21 120280 yes atrial fibrillation chr1 GJA5 1q21 121013 yes Thrombocytopenia absent radius syndrome chr1 GJA8 1q21 600897 yes Short stature chr1 LHX4 1q25 602146 yes Van der Woude syndrome chr1 IRF6 1q32 607199 yes Fryns 1q41 chr1 DISP1 1q41 607502 yes autism chr1 DISC1 1q42 605210 yes MR chr1 TBCE 1q42 604934 yes Feingold chr2 MYCN 2q24 164840 yes Pseudovaginal perineoscrotal hypospadias chr2 SRD5A2 2p23 607306 yes Noonan 4 chr2 SOS1 2p22 182530 yes Cystinuria with mitochondrial disease chr2 SLC3A1 2p21 104614 yes Cystinuria with mitochondrial disease chr2 PREPL 2p21 104614 yes Holopresencaphly 2 chr2 SIX3 2p21 603714 yes autism chr2 NRXN1 2p16 600565 yes MicrodeletionReg 2p15p16.1 microdeletion chr2 2p15 602559 yes ion Nephronophthsis 1 chr2 NPHP1 2q13 607100 yes Holoprosencephaly 9 chr2 GLI2 2q14 165230 yes visceral heterotaxy chr2 CFC1 2q21 605194 yes Mowat-Wilson syndrome chr2 ZEB2 2q22 605802 yes autism chr2 SLC4A10 2q24 605556 yes SCN1A-related seizures chr2 SCN1A 2q24 182389 yes HYPOMYELINATION, GLOBAL CEREBRAL chr2 SLC25A12 2q31 603667 yes Split/hand foot malformation -5 chr2 DLX1 2q31 600029 yes