Characterization of Alginate Lyase from Microbulbifer Mangrovi Sp. Nov
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

MS#6 (Tayco Et Al)
Philippine Journal of Science 142 (1): 45-54, June 2013 ISSN 0031 - 7683 Date Received: ?? Feb 20?? Characterization of a κ-Carrageenase-producing Marine Bacterium, Isolate ALAB-001 Crimson C. Tayco1, Francis A. Tablizo1, Raymond S. Regalia2 and Arturo O. Lluisma1* 1The Marine Science Institute, University of the Philippines Diliman, Quezon City, Philippines 1101 2Center for Marine Bio-Innovation, School of Biotechnology and Biomolecular Sciences, Faculty of Science, The University of New South Wales, Sydney, Australia 2052 Carrageenases are glycoside hydrolases that specifically degrade carrageenan, a highly anionic polysaccharide found in the cell wall of many red algal species. To date, only a few of these enzymes have been characterized, and identifying additional sources is important considering the role of carrageenases in production of carrageenan derivatives. In this paper, we report the characterization of a marine bacterial strain that produces κ-carrageenase. The strain, which we designate as ALAB-001, was isolated from diseased thallus fragments of the red alga Kappaphycus alvarezii, a commercially important source of carrageenan. Genotypic and phenotypic data suggest that the isolate belongs to a relatively poorly-characterized group of bacteria in Alteromonadaceae (Alteromonadales) and is closely related to Marinimicrobium and Microbulbifer. Significant κ-carrageenase activity (175 U/mL) was evident when the isolate was grown in the presence of κ-carrageenan. Activity against starch was also high (180 U/mL), but activity against agar, alginate, cellulose, ι-carrageenan, and λ-carrageenan was significantly lower (25-50 U/mL). Laboratory-scale production of the enzyme using batch cultures of the isolate was achieved by optimizing culture medium, length of culture time and degree temperature. -

View a Copy of This Licence, Visit
Raimundo et al. Microbiome (2021) 9:43 https://doi.org/10.1186/s40168-020-00970-2 RESEARCH Open Access Functional metagenomics reveals differential chitin degradation and utilization features across free-living and host-associated marine microbiomes I. Raimundo1†, R. Silva1†, L. Meunier1,2, S. M. Valente1, A. Lago-Lestón3, T. Keller-Costa1* and R. Costa1,4,5,6* Abstract Background: Chitin ranks as the most abundant polysaccharide in the oceans yet knowledge of shifts in structure and diversity of chitin-degrading communities across marine niches is scarce. Here, we integrate cultivation- dependent and -independent approaches to shed light on the chitin processing potential within the microbiomes of marine sponges, octocorals, sediments, and seawater. Results: We found that cultivatable host-associated bacteria in the genera Aquimarina, Enterovibrio, Microbulbifer, Pseudoalteromonas, Shewanella, and Vibrio were able to degrade colloidal chitin in vitro. Congruent with enzymatic activity bioassays, genome-wide inspection of cultivated symbionts revealed that Vibrio and Aquimarina species, particularly, possess several endo- and exo-chitinase-encoding genes underlying their ability to cleave the large chitin polymer into oligomers and dimers. Conversely, Alphaproteobacteria species were found to specialize in the utilization of the chitin monomer N-acetylglucosamine more often. Phylogenetic assessments uncovered a high degree of within-genome diversification of multiple, full-length endo-chitinase genes for Aquimarina and Vibrio strains, suggestive of a versatile chitin catabolism aptitude. We then analyzed the abundance distributions of chitin metabolism-related genes across 30 Illumina-sequenced microbial metagenomes and found that the endosymbiotic consortium of Spongia officinalis is enriched in polysaccharide deacetylases, suggesting the ability of the marine sponge microbiome to convert chitin into its deacetylated—and biotechnologically versatile—form chitosan. -

The Microbiome of North Sea Copepods
Helgol Mar Res (2013) 67:757–773 DOI 10.1007/s10152-013-0361-4 ORIGINAL ARTICLE The microbiome of North Sea copepods G. Gerdts • P. Brandt • K. Kreisel • M. Boersma • K. L. Schoo • A. Wichels Received: 5 March 2013 / Accepted: 29 May 2013 / Published online: 29 June 2013 Ó Springer-Verlag Berlin Heidelberg and AWI 2013 Abstract Copepods can be associated with different kinds Keywords Bacterial community Á Copepod Á and different numbers of bacteria. This was already shown in Helgoland roads Á North Sea the past with culture-dependent microbial methods or microscopy and more recently by using molecular tools. In our present study, we investigated the bacterial community Introduction of four frequently occurring copepod species, Acartia sp., Temora longicornis, Centropages sp. and Calanus helgo- Marine copepods may constitute up to 80 % of the meso- landicus from Helgoland Roads (North Sea) over a period of zooplankton biomass (Verity and Smetacek 1996). They are 2 years using DGGE (denaturing gradient gel electrophore- key components of the food web as grazers of primary pro- sis) and subsequent sequencing of 16S-rDNA fragments. To duction and as food for higher trophic levels, such as fish complement the PCR-DGGE analyses, clone libraries of (Cushing 1989; Møller and Nielsen 2001). Copepods con- copepod samples from June 2007 to 208 were generated. tribute to the microbial loop (Azam et al. 1983) due to Based on the DGGE banding patterns of the two years sur- ‘‘sloppy feeding’’ (Møller and Nielsen 2001) and the release vey, we found no significant differences between the com- of nutrients and DOM from faecal pellets (Hasegawa et al. -

Study of Bacterial Communities in Mussel Mytilus Galloprovincialis (Bivalvia: Mytilidae) by a Combination of 16S Crdna and 16S Rdna Sequencing
Central JSM Microbiology Research Article Corresponding author Simone Cappello, Istituto per l’Ambiente Marino Costiero (IAMC) – CNR of Messina, Spianata San Raineri, Study of Bacterial 86 - 98122 Messina, Italy; Tel: +39-090-6015421; Fax: +39- 090-669003, Email: Submitted: 28 October 2014 Communities in Mussel Mytilus Accepted: 09 January 2015 Published: 12 January 2015 galloprovincialis (Bivalvia: Copyright © 2015 Cappello et al. Mytilidae) by a Combination OPEN ACCESS Keywords of 16s Crdna and 16s Rdna • Mytilus galloprovincialis • Microbial community Sequencing • Symbiont Simone Cappello1*, Anna Volta1,2, Santina Santisi1,3, Lucrezia Genovese1, Giulia Maricchiolo1, Martina Bonsignore1 and Michail M. Yakimov1 1Istituto per l’Ambiente Marino Costiero (IAMC) – CNR of Messina, Italy 2Department of Industrial and Mechanical Engineering, University of Catania, Italy 3PhD School of “Biology and Cellular Biotechnology” of University of Messina, Italy Abstract In this study has been analyzed the genetic potential (rDNA) versus expression (crDNA) of microbial populations associated to gills of living mussel Mytilus galloprovincialis (Bivalvia: Mytilidae) in natural environment. Data obtained (16S rDNA/crDNA clones libraries) showed as sequences mainly related to Bacteroides/ Chlorobi, Firmicutes and Gamma-Proteobacteria groups are specific in live mussels. It is presumed that further studies of microbial population structure with culture-independent methods will demonstrate the active interactions (symbiosis) between filter-feeding organisms -

Supplement of Biogeosciences, 13, 5527–5539, 2016 Doi:10.5194/Bg-13-5527-2016-Supplement © Author(S) 2016
Supplement of Biogeosciences, 13, 5527–5539, 2016 http://www.biogeosciences.net/13/5527/2016/ doi:10.5194/bg-13-5527-2016-supplement © Author(s) 2016. CC Attribution 3.0 License. Supplement of Seasonal changes in the D / H ratio of fatty acids of pelagic microorganisms in the coastal North Sea Sandra Mariam Heinzelmann et al. Correspondence to: Sandra Mariam Heinzelmann ([email protected]) The copyright of individual parts of the supplement might differ from the CC-BY 3.0 licence. Figure legends Supplementary Figure S1 Phylogenetic tree of 16S rRNA gene sequence reads assigned to Bacteroidetes. Scale bar indicates 0.10 % estimated sequence divergence. Groups containing sequences are highlighted. Figure S2 Phylogenetic tree of 16S rRNA gene sequence reads assigned to Alphaproteobacteria. Scale bar indicates 0.10 % estimated sequence divergence. Groups containing sequences are highlighted. Figure S3 Phylogenetic tree of 16S rRNA gene sequence reads assigned to Gammaproteobacteria. Scale bar indicates 0.10 % estimated sequence divergence. Groups containing sequences are highlighted. Figure S4 δDwater versus salinity of North Sea SPM sampled in 2013. Bacteroidetes figS01 group including Prevotellaceae Bacteroidaceae_Bacteroides RH-aaj90h05 RF16 S24-7 gir-aah93ho Porphyromonadaceae_1 ratAN060301C Porphyromonadaceae_2 3M1PL1-52 termite group Porphyromonadaceae_Paludibacter EU460988, uncultured bacterium, red kangaroo feces Porphyromonadaceae_3 009E01-B-SD-P15 Rikenellaceae MgMjR-022 BS11 gut group Rs-E47 termite group group including termite group FTLpost3 ML635J-40 aquatic group group including gut group vadinHA21 LKC2.127-25 Marinilabiaceae Porphyromonadaceae_4 Sphingobacteriia_Sphingobacteriales_1 group including Cytophagales Bacteroidetes Incertae Sedis_Unknown Order_Unknown Family_Prolixibacter WCHB1-32 SB-1 vadinHA17 SB-5 BD2-2 Ika-33 VC2.1 Bac22 Flavobacteria_Flavobacteriales including e.g. -

Bacterial Taxa Based on Greengenes Database GS1A PS1B ABY1 OD1
A1: Bacterial taxa based on GreenGenes database GS1A PS1B ABY1_OD1 0.1682 0.024 Bacteria;ABY1_OD1;ABY1_OD1_unclassified 1 0 Bacteria;ABY1_OD1;FW129;FW129_unclassified 4 0 Bacteria;ABY1_OD1;FW129;KNA6-NB12;KNA6-NB12_unclassified 5 0 Bacteria;ABY1_OD1;FW129;KNA6-NB29;KNA6-NB29_unclassified 0 1 Acidobacteria 0.7907 4.509 Bacteria;Acidobacteria;Acidobacteria_unclassified 4 31 Bacteria;Acidobacteria;Acidobacteria-5;Acidobacteria-5_unclassified 0 1 Bacteria;Acidobacteria;BPC015;BPC015_unclassified 8 30 Bacteria;Acidobacteria;BPC102;BPC102_unclassified 9 43 Bacteria;Acidobacteria;Chloracidobacteria;Ellin6075;Ellin6075_unclassified 1 0 Bacteria;Acidobacteria;iii1-15;Acidobacteria-6;RB40;RB40_unclassified 0 5 Bacteria;Acidobacteria;iii1-15;iii1-15_unclassified 1 8 Bacteria;Acidobacteria;iii1-15;Riz6I;Unclassified 0 1 Bacteria;Acidobacteria;iii1-8;Unclassified 0 2 Bacteria;Acidobacteria;OS-K;OS-K_unclassified 18 17 Bacteria;Acidobacteria;RB25;RB25_unclassified 6 47 Bacteria;Acidobacteria;Solibacteres;Solibacteres_unclassified 0 1 Actinobacteria 2.1198 6.642 Bacteria;Actinobacteria;Acidimicrobidae;Acidimicrobidae_unclassified 10 70 Bacteria;Actinobacteria;Acidimicrobidae;CL500-29;ML316M-15;ML316M-15_unclassified 0 3 Bacteria;Actinobacteria;Acidimicrobidae;EB1017_group;Acidimicrobidae_bacterium_Ellin7143;Unclassified 6 1 Bacteria;Actinobacteria;Acidimicrobidae;koll13;JTB31;BD2-10;BD2-10_unclassified 1 5 Bacteria;Actinobacteria;Acidimicrobidae;koll13;JTB31;Unclassified 16 37 Bacteria;Actinobacteria;Acidimicrobidae;koll13;koll13_unclassified 81 25 Bacteria;Actinobacteria;Acidimicrobidae;Microthrixineae;Microthrixineae_unclassified -

Hydrocarbon Pollutants Shape Bacterial Community Assembly of Harbor Sediments
Hydrocarbon pollutants shape bacterial community assembly of harbor sediments Item Type Article Authors Barbato, Marta; Mapelli, Francesca; Magagnini, Mirko; Chouaia, Bessem; Armeni, Monica; Marasco, Ramona; Crotti, Elena; Daffonchio, Daniele; Borin, Sara Citation Hydrocarbon pollutants shape bacterial community assembly of harbor sediments 2016 Marine Pollution Bulletin Eprint version Post-print DOI 10.1016/j.marpolbul.2016.01.029 Publisher Elsevier BV Journal Marine Pollution Bulletin Rights NOTICE: this is the author’s version of a work that was accepted for publication in Marine Pollution Bulletin. Changes resulting from the publishing process, such as peer review, editing, corrections, structural formatting, and other quality control mechanisms may not be reflected in this document. Changes may have been made to this work since it was submitted for publication. A definitive version was subsequently published in Marine Pollution Bulletin, 2 February 2016. DOI: 10.1016/ j.marpolbul.2016.01.029 Download date 25/09/2021 06:13:34 Link to Item http://hdl.handle.net/10754/597022 Hydrocarbon pollutants shape bacterial community assembly of harbor sediments Marta Barbato1§, Francesca Mapelli1§, Mirko Magagnini2, Bessem Chouaia1,a, Monica Armeni2, Ramona Marasco3, Elena Crotti1, Daniele Daffonchio1,3, Sara Borin1* 1Department of Food, Environmental and Nutritional Sciences (DeFENS), University of Milan, Milan, Italy 2 EcoTechSystems Ltd., Ancona, Italy. 3 Biological and Environmental Sciences and Engineering Division (BESE). King Abdullah -

Melitea Salexigens Gen. Nov., Sp. Nov., a Gammaproteobacterium from The
Melitea salexigens gen. nov., sp. nov., a gammaproteobacterium from the Mediterranean Sea Laurent Urios, Hélène Agogué, Laurent Intertaglia, Françoise Lesongeur, Philippe Lebaron To cite this version: Laurent Urios, Hélène Agogué, Laurent Intertaglia, Françoise Lesongeur, Philippe Lebaron. Melitea salexigens gen. nov., sp. nov., a gammaproteobacterium from the Mediterranean Sea. International Journal of Systematic and Evolutionary Microbiology, Microbiology Society, 2008, 58, pp.2489-2483. 10.1099/ijs.0.65685-0. hal-01102906 HAL Id: hal-01102906 https://hal.archives-ouvertes.fr/hal-01102906 Submitted on 13 Jan 2015 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. International Journal of Systematic and Evolutionary Microbiology (2008), 58, 2479–2483 DOI 10.1099/ijs.0.65685-0 Melitea salexigens gen. nov., sp. nov., a gammaproteobacterium from the Mediterranean Sea Laurent Urios,1,2 He´le`ne Agogue´,1,2 Laurent Intertaglia,1,2 Franc¸oise Lesongeur3 and Philippe Lebaron1,2 Correspondence 1Universite´ Pierre et Marie Curie – Paris 6, Laboratoire ARAGO, Avenue du Fontaule´, BP 44, F- Philippe Lebaron 66650 Banyuls-sur-Mer, France [email protected] 2CNRS, UMR7621, Laboratoire d’Oce´anographie Biologique de Banyuls, Avenue du Fontaule´,BP 44, F-66650 Banyuls-sur-Mer, France 3Laboratoire de Microbiologie des Environnements Extreˆmes, UMR 6197, IFREMER, Centre de Brest, BP 70, F-29280 Plouzane´, France A novel aerobic, Gram-negative bacterial strain, designated 5IX/A01/131T, was isolated from waters in the coastal north-western Mediterranean Sea. -

Resource for Scientific Discovery
RESOU R CE OPEN 1,003 reference genomes of bacterial and archaeal isolates expand coverage of the tree of life Supratim Mukherjee1,10, Rekha Seshadri1,10, Neha J Varghese1, Emiley A Eloe-Fadrosh1, Jan P Meier-Kolthoff2 , Markus Göker2 , R Cameron Coates1,9, Michalis Hadjithomas1, Georgios A Pavlopoulos1 , David Paez-Espino1 , Yasuo Yoshikuni1, Axel Visel1 , William B Whitman3, George M Garrity4,5, Jonathan A Eisen6, Philip Hugenholtz7 , Amrita Pati1,9, Natalia N Ivanova1, Tanja Woyke1, Hans-Peter Klenk8 & Nikos C Kyrpides1 We present 1,003 reference genomes that were sequenced as part of the Genomic Encyclopedia of Bacteria and Archaea (GEBA) initiative, selected to maximize sequence coverage of phylogenetic space. These genomes double the number of existing type strains and expand their overall phylogenetic diversity by 25%. Comparative analyses with previously available finished and draft genomes reveal a 10.5% increase in novel protein families as a function of phylogenetic diversity. The GEBA genomes recruit 25 million previously unassigned metagenomic proteins from 4,650 samples, improving their phylogenetic and functional interpretation. We identify numerous biosynthetic clusters and experimentally validate a divergent phenazine cluster with potential new chemical structure and antimicrobial activity. This Resource is the largest single release of reference genomes to date. Bacterial and archaeal isolate sequence space is still far from saturated, and future endeavors in this direction will continue to be a valuable resource for scientific discovery. Systematic surveys of the diversity of cultivated microorganisms have subsequent experiments. Typically, a type strain has well-character- lagged behind improvements in sequencing technologies. Traditionally, ized taxonomic and phenotypic data, isolation source metadata, and most isolate sequencing projects are chosen based on the clinical or other criteria, as defined by the International Code of Nomenclature biotechnological relevance of the target organisms or their physiology1. -

Taxogenomic and Metabolic Insights Into Marinobacterium Ramblicola Sp
microorganisms Article Taxogenomic and Metabolic Insights into Marinobacterium ramblicola sp. nov., a New Slightly Halophilic Bacterium Isolated from Rambla Salada, Murcia Ana Durán-Viseras 1,2,* , David J. Castro 1,2, José Carlos Reina 1,2 , Victoria Béjar 1,2 and Fernando Martínez-Checa 1,2,* 1 Microbial Exopolysaccharide Research Group, Department of Microbiology, Pharmacy Faculty, Campus de Cartuja s.n., 18071 Granada, Spain; [email protected] (D.J.C.); [email protected] (J.C.R.); [email protected] (V.B.) 2 Biomedical Research Center, Institute of Biotechnology, 18016 Granada, Spain * Correspondence: [email protected] (A.D.-V.); [email protected] (F.M.-C.) Abstract: A Gram-negative, motile, rod-shaped bacteria, designated D7T, was isolated by using the dilution-to-extinction method, from a soil sample taken from Rambla Salada (Murcia, Spain). Growth of strain D7T was observed at 15–40 ◦C (optimum, 37 ◦C), pH 5–9 (optimum, 7) and 0–7.5% (w/v) NaCl (optimum, 3%). It is facultatively anaerobic. Phylogenetic analysis based on 16S rRNA gene sequence showed it belongs to the genus Marinobacterium. The in silico DDH and ANI against closest Marinobacterium relatives support its placement as a new species within this genus. The major fatty T acids of strain D7 were C16:0, summed feature 3 (C16:1 w7c/C16:1 w6c) and summed feature 8 (C18:1 Citation: Durán-Viseras, A.; Castro, w7c/C18:1 w6c). The polar lipid profile consists of phosphatidylethanolamine, phosphatidylglycerol and D.J.; Reina, J.C.; Béjar, V.; two uncharacterized lipids. Ubiquinone 8 was the unique isoprenoid quinone detected. -

The Marine Air-Water, Located Between the Atmosphere and The
A survey on bacteria inhabiting the sea surface microlayer of coastal ecosystems Hélène Agoguéa, Emilio O. Casamayora,b, Muriel Bourrainc, Ingrid Obernosterera, Fabien Jouxa, Gerhard Herndld and Philippe Lebarona aObservatoire Océanologique, Université Pierre et Marie Curie, UMR 7621-INSU-CNRS, BP44, 66651 Banyuls-sur-Mer Cedex, France bUnidad de Limnologia, Centro de Estudios Avanzados de Blanes-CSIC. Acc. Cala Sant Francesc, 14. E-17300 Blanes, Spain cCentre de Recherche Dermatologique Pierre Fabre, BP 74, 31322, Castanet Tolosan, France dDepartment of Biological Oceanography, Royal Institute for Sea Research (NIOZ), P.O. Box 59, 1790 AB Den Burg, The Netherlands 1 Summary Bacterial populations inhabiting the sea surface microlayer from two contrasted Mediterranean coastal stations (polluted vs. oligotrophic) were examined by culturig and genetic fingerprinting methods and were compared with those of underlying waters (50 cm depth), for a period of two years. More than 30 samples were examined and 487 strains were isolated and screened. Proteobacteria were consistently more abundant in the collection from the pristine environment whereas Gram-positive bacteria (i.e., Actinobacteria and Firmicutes) were more abundant in the polluted site. Cythophaga-Flavobacter–Bacteroides (CFB) ranged from 8% to 16% of total strains. Overall, 22.5% of the strains showed a 16S rRNA gene sequence similarity only at the genus level with previously reported bacterial species and around 10.5% of the strains showed similarities in 16S rRNA sequence below 93% with reported species. The CFB group contained the highest proportion of unknown species, but these also included Alpha- and Gammaproteobacteria. Such low similarity values showed that we were able to culture new marine genera and possibly new families, indicating that the sea-surface layer is a poorly understood microbial environment and may represent a natural source of new microorganisms. -
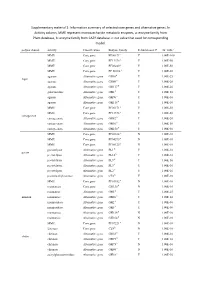
Information Summary of Selected Core Genes and Alternative Genes. in Activity Column, MME Represent Mo
Supplementary material 1: Information summary of selected core genes and alternative genes. In Activity column, MME represent monosaccharide metabolic enzymes, a: enzyme family from Pfam database, b: enzyme family from CAZY database. c: cut value that used for corresponding model. polysaccharide activity Classification Enzyme family Rebuild model? cut_valuec MME Core gene PF00171a Y 1.00E-100 MME Core gene PF13378 a Y 1.00E-50 MME Core gene PF08240 a Y 1.00E-50 MME Core gene PF 00106 a Y 1.00E-50 agarase Alternative gene GH16b Y 1.00E-25 Agar agarase Alternative gene GH86 b Y 1.00E-20 agarase Alternative gene GH117 b Y 1.00E-20 galactosidase Alternative gene GH2 b Y 1.00E-50 agarase Alternative gene GH96 b Y 1.00E-10 agarase Alternative gene GH118 b Y 1.00E-10 MME Core gene PF00171 a Y 1.00E-50 MME Core gene PF13378 a Y 1.00E-50 carrageenan carrageenase Alternative gene GH82 b Y 1.00E-20 carrageenase Alternative gene GH16 b Y 1.00E-50 carrageenase Alternative gene GH150 b Y 1.00E-50 MME Core gene PF02614 a N 1.00E-10 MME Core gene PF04295 a N 1.00E-10 MME Core gene PF08125 a N 1.00E-10 pectatelyase Alternative gene PL1 b Y 1.00E-10 pectin pectatelyase Alternative gene PL10 b Y 1.00E-10 pectatelyase Alternative gene PL9 b Y 1.00E-50 pectatelyase Alternative gene PL3 b Y 1.00E-10 pectatelyase Alternative gene PL2 b Y 1.00E-10 pectinmethylesterase Alternative gene CE8 b Y 1.00E-10 MME Core gene PF01182 a N 1.00E-10 mannanase Core gene GH130 b N 1.00E-10 mannanase Alternative gene GH5 b Y 1.00E-25 mannan mannanase Alternative gene GH26 b