Molecular Phylogenetics of Melastomataceae and Memecylaceae: Implications for Character Evolution1
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Kilaka Forest
Kilaka Forest Conservation Area Management Plan Copyright: © 2016 Wildlife Conservation Society Reproduction of this publication for educational or other non-commercial purposes is authorized without prior written permission from the copyright holder provided that the source is fully acknowledged. Reproduction of this publication for resale or other commercial purposes is prohibited withoutprior written consent of the copyright owner. Citation: WCS (2016) Kilaka Forest Conservation Area Management Plan. Wildlife Conservation Society, Suva, Fiji. 34 pp. Photograph (front cover): ©Ruci Lumelume/WCS Graphic design & Layout: cChange NOTE: This management plan may be amended from time to time. To obtain a copy of the current management plan, please contact: Wildlife Conservation Society Fiji Country Program 11 Ma’afu Street Suva Republic of Fiji Islands Telephone: +679 331 5174 Email: [email protected] Kilaka Forest Conservation Area Management Committee Kilaka Village Kubulau District Bua Province Republic of Fiji Kubulau Resource Management Committee Kubulau District Bua Province Republic of Fiji ENDORSEMENT On this day, 24 November, 2016 at Kilaka Village in the district of Kubulau, Bua Province, Vanua Levu in the Republic of Fiji Islands, we the undersigned endorse this management plan and its implementation. We urge the people of all communities in Kubulau and key stakeholders from government, private and non-government sectors to observe the plan and make every effort to ensure effective implementation. Minister, Ministry of Forests Tui -

Survey of Roadside Alien Plants in Hawai`I Volcanoes National Park and Adjacent Residential Areas 2001–2005
Technical Report HCSU-032 SURVEY OF ROADSIDE ALIEN PLANts IN HAWAI`I VOLCANOES NATIONAL PARK AND ADJACENT RESIDENTIAL AREAS 2001–2005 Linda W. Pratt1 Keali`i F. Bio2 James D. Jacobi1 1 U.S. Geological Survey, Pacific Island Ecosystems Research Center, Kilauea Field Station, P.O. Box 44, Hawaii National Park, HI 96718 2 Hawai‘i Cooperative Studies Unit, University of Hawai‘i at Hilo, P.O. Box 44, Hawai‘i National Park, HI 96718 Hawai‘i Cooperative Studies Unit University of Hawai‘i at Hilo 200 W. Kawili St. Hilo, HI 96720 (808) 933-0706 September 2012 This product was prepared under Cooperative Agreement CA03WRAG0036 for the Pacific Island Ecosystems Research Center of the U.S. Geological Survey. Technical Report HCSU-032 SURVEY OF ROADSIDE ALIEN PLANTS IN HAWAI`I VOLCANOES NATIONAL PARK AND ADJACENT RESIDENTIAL AREAS 2001–2005 1 2 1 LINDA W. PRATT , KEALI`I F. BIO , AND JAMES D. JACOBI 1 U.S. Geological Survey, Pacific Island Ecosystems Research Center, Kīlauea Field Station, P.O. Box 44, Hawai`i Volcanoes National Park, HI 96718 2 Hawaii Cooperative Studies Unit, University of Hawai`i at Hilo, Hilo, HI 96720 Hawai`i Cooperative Studies Unit University of Hawai`i at Hilo 200 W. Kawili St. Hilo, HI 96720 (808) 933-0706 September 2012 This article has been peer reviewed and approved for publication consistent with USGS Fundamental Science Practices ( http://pubs.usgs.gov/circ/1367/ ). Any use of trade, firm, or product names is for descriptive purposes only and does not imply endorsement by the U.S. Government. -

Phylogeny and Classification of the Melastomataceae and Memecylaceae
Nord. J. Bot. - Section of tropical taxonomy Phylogeny and classification of the Melastomataceae and Memecy laceae Susanne S. Renner Renner, S. S. 1993. Phylogeny and classification of the Melastomataceae and Memecy- laceae. - Nord. J. Bot. 13: 519-540. Copenhagen. ISSN 0107-055X. A systematic analysis of the Melastomataceae, a pantropical family of about 4200- 4500 species in c. 166 genera, and their traditional allies, the Memecylaceae, with c. 430 species in six genera, suggests a phylogeny in which there are two major lineages in the Melastomataceae and a clearly distinct Memecylaceae. Melastomataceae have close affinities with Crypteroniaceae and Lythraceae, while Memecylaceae seem closer to Myrtaceae, all of which were considered as possible outgroups, but sister group relationships in this plexus could not be resolved. Based on an analysis of all morph- ological and anatomical characters useful for higher level grouping in the Melastoma- taceae and Memecylaceae a cladistic analysis of the evolutionary relationships of the tribes of the Melastomataceae was performed, employing part of the ingroup as outgroup. Using 7 of the 21 characters scored for all genera, the maximum parsimony program PAUP in an exhaustive search found four 8-step trees with a consistency index of 0.86. Because of the limited number of characters used and the uncertain monophyly of some of the tribes, however, all presented phylogenetic hypotheses are weak. A synapomorphy of the Memecylaceae is the presence of a dorsal terpenoid-producing connective gland, a synapomorphy of the Melastomataceae is the perfectly acrodro- mous leaf venation. Within the Melastomataceae, a basal monophyletic group consists of the Kibessioideae (Prernandra) characterized by fiber tracheids, radially and axially included phloem, and median-parietal placentation (placentas along the mid-veins of the locule walls). -

Wood Anatomy of Lythraceae Assigned to The
Ada Bot. Neerl. 28 (2/3), May 1979, p.117-155. Wood anatomy of the Lythraceae P. Baas and R.C.V.J. Zweypfenning Rijksherbarium, Leiden, The Netherlands SUMMARY The wood anatomy of 18 genera belonging to the Lythraceae is described. The diversity in wood structure of extant Lythraceae is hypothesized to be derived from a prototype with scanty para- I tracheal parenchyma, heterogeneous uniseriate and multiseriate rays, (septate)libriform fibres with minutely bordered pits, and vessels with simple perforations. These characters still prevail in a number of has been limited in of Lythraceae. Specialization very most Lythraceae shrubby or herbaceous habit: these have juvenilistic rays composed mainly of erect rays and sometimes com- pletely lack axial parenchyma. Ray specialization towards predominantly uniseriate homogeneous concomitant with fibre abundant and with rays, dimorphism leading to parenchyma differentiation, the advent of chambered crystalliferous fibres has been traced in the “series” Ginoria , Pehria, Lawsonia , Physocalymma and Lagerstroemia. The latter genus has the most specialized wood anatomy in the family and has species with abundant parenchyma aswell as species with alternating fibres. with its bands of dimorphous septate Pemphis represents an independent specialization vasicentric parenchyma and thick-walled nonseptate fibres. The affinities of with other are discussed. Pun Psiloxylon, Lythraceae Myrtales ica, Rhynchocalyx , Oliniaceae,Alzatea, Sonneratiaceae, Onagraceae and Melastomataceae all resemble Lythraceae in former accommodated in the without their wood anatomy. The three genera could even be family its wood anatomical Alzatea and Sonneratia differ in minor details extending range. Oliniaceae, only from order facilitate identification of wood tentative the Lythraceae. In to samples, keys to genera or groups ofgenera of Lythraceae as well as to some species of Lagerstroemiaare presented. -

Vitales, C Nymphaeales Austrobaileyales
Amborellales Vitales, C Nymphaeales Austrobaileyales Acorales G Eenzaadlobbigen G Alismatales Vitales Petrosaviales Pandanales De Wijnstokfamilie (Vitaceae), d Dioscoreales vroeger samen met de Wegedoo Liliales geplaatst, omdat in beide famil Asparagales kroonbladen staan. Dat de Vital Arecales maar niet precies waar. Hiervoo G Commeliniden G Dasypogonales Poales Commelinales Crossosomatales Zingiberales Deze nieuwe orde in de Rosiden ordes en wordt ondersteund do Ceratophyllales kenmerken, zoals de structuur v Chloranthales afkomstig uit de Violales, Celast Het zijn 5 kleine families uit wa Canellales Piperales Aphloiaceae, Geissolomataceae G Magnoliiden G Magnoliales Stachyuraceae, en 2 iets grotere Laurales (Staphyleacea) en de Crossosom Ranunculales Sabiales Proteales Vitaceae Trochodendrales Buxales Aphloiacea Geissoloma Gunnerales Ixerbaceae Berberidopsidales Strasburge Dilleniales Staphyleac Caryophyllales Stachyurac Santalales Crossosom Saxifragales Melianthac G Geavanceerde tweezaadlobbigen G Vitales Francoacea Crossosomatales Ledocarpa Geraniales Vivianiacea Myrtales Geraniacea Zygophyllales Combretac Celastrales Lythraceae Malpighiales Onagracea G Fabiden G Oxalidales Vochysiace Fabales Myrtaceae G Rosiden G Rosales Crypteroni Cucurbitales Alzateacea Fagales Rhynchoca Oliniaceae Brassicales Penaeacea G G Malviden Malvales Melastoma Sapindales Cornales kenmerken. Uit de Polygalales z Ericales G Asteriden G erbij gevoegd, een kleine famili Garryales Afrika en Amerika. G Lamiiden G Gentianales De bladeren zijn meestal tegeno Solanales -

Systematics and Relationships of Tryssophyton (Melastomataceae
A peer-reviewed open-access journal PhytoKeys 136: 1–21 (2019)Systematics and relationships of Tryssophyton (Melastomataceae) 1 doi: 10.3897/phytokeys.136.38558 RESEARCH ARTICLE http://phytokeys.pensoft.net Launched to accelerate biodiversity research Systematics and relationships of Tryssophyton (Melastomataceae), with a second species from the Pakaraima Mountains of Guyana Kenneth J. Wurdack1, Fabián A. Michelangeli2 1 Department of Botany, MRC-166 National Museum of Natural History, Smithsonian Institution, P.O. Box 37012, Washington, DC 20013-7012, USA 2 The New York Botanical Garden, 2900 Southern Blvd., Bronx, NY 10458, USA Corresponding author: Kenneth J. Wurdack ([email protected]) Academic editor: Ricardo Kriebel | Received 25 July 2019 | Accepted 30 October 2019 | Published 10 December 2019 Citation: Wurdack KJ, Michelangeli FA (2019) Systematics and relationships of Tryssophyton (Melastomataceae), with a second species from the Pakaraima Mountains of Guyana. PhytoKeys 136: 1–21. https://doi.org/10.3897/ phytokeys.136.38558 Abstract The systematics of Tryssophyton, herbs endemic to the Pakaraima Mountains of western Guyana, is re- viewed and Tryssophyton quadrifolius K.Wurdack & Michelang., sp. nov. from the summit of Kamakusa Mountain is described as the second species in the genus. The new species is distinguished from its closest relative, Tryssophyton merumense, by striking vegetative differences, including number of leaves per stem and leaf architecture. A phylogenetic analysis of sequence data from three plastid loci and Melastomata- ceae-wide taxon sampling is presented. The two species of Tryssophyton are recovered as monophyletic and associated with mostly Old World tribe Sonerileae. Fruit, seed and leaf morphology are described for the first time, biogeography is discussed and both species are illustrated. -

The Uses of Molecular Dating Analyses in Evolutionary Studies, with Examples from the Angiosperms
Zurich Open Repository and Archive University of Zurich Main Library Strickhofstrasse 39 CH-8057 Zurich www.zora.uzh.ch Year: 2006 The uses of molecular dating analyses in evolutionary studies, with examples from the angiosperms Rutschmann, Frank Kaspar Abstract: Die sechs Kapitel der vorliegenden Dissertation befassen sich alle mit verschiedenen Aspekten von molekularen Datierungsmethoden, englisch molecular dating methods genannt. Dabei reicht die Spannweite der behandelten Themen von experimentellen Untersuchungen der Methoden selbst bis hin zur praktischen Anwendung der molekularen Altersbestimmung für die Aufklärung von biologischen und evolutionsgeschichtlichen Fragen bei verschiedenen Gruppen von Blütenpflanzen (Angiospermen). The six chapters that compose this dissertation are all related to various aspects of molecular dating, ranging from more methodological and experimental work to the application of different methods in the context of biological and evolutionary questions and hypotheses related to different groups of flowering plants (Angiosperms). Posted at the Zurich Open Repository and Archive, University of Zurich ZORA URL: https://doi.org/10.5167/uzh-163459 Dissertation Published Version Originally published at: Rutschmann, Frank Kaspar. The uses of molecular dating analyses in evolutionary studies, with examples from the angiosperms. 2006, University of Zurich, Faculty of Science. The Uses of Molecular Dating Analyses in Evolutionary Studies, with Examples from the Angiosperms Dissertation zur Erlangung der naturwissenschaftlichen Doktorwürde (Dr. sc. nat.) vorgelegt der Mathematisch-naturwissenschaftlichen Fakultt der niversitt Zürich von Frank Kaspar Rutschmann von Zürich ZH Promotionskomitee: Prof. Dr. Elena Conti ()orsitz) Prof. Dr. Peter Linder Dr. Torsten Eriksson Zürich 200. Acknowledgements First of all, I would like to express my gratitude towards my supervisor, Elena Conti. -

9 Costion Plant Endemism 133-166 PROOFS
Micronesica 41(1): 131–164, 2009 Plant Endemism, Rarity, and Threat in Palau, Micronesia: A Geographical Checklist and Preliminary Red List Assessment 1 CRAIG M. COSTION Department of Ecology and Evolutionary Biology, School of Earth and Environmental Sciences, University of Adelaide, Adelaide SA 5001 [email protected] ANN HILLMANN KITALONG The Environment, Inc., P.O. Box 1696, Koror, Palau 96940 TARITA HOLM Palau Conservation Society/PALARIS, P.O. Box 1811, Koror, Palau, 96940 Abstract—An official checklist of the endemic plant species of Palau has been long awaited, and is presented here for the first time. For each species a substrate limitation, growth form, and relative abundance is listed. In addition an IUCN red list assessment was conducted using all available data. For over half of the endemic species there is insufficient data to provide a red listing status however an expected minimum number of threatened plants out of the total is inferred. Approximately 15% of Palau’s endemic plants are believed to be only known from the type collection and many more only known from a few collections. These taxa however may now be prioritized and targeted for future inventory and research. The taxonomic robustness of several of these taxa is questionable and it is expected that more endemic species will be lost to synonymy in the future. Previous estimations have significantly over-estimated the rate of plant endemism in Palau (e.g., 194). Here, 130 plants are recognized for Palau, making its level of plant endem- ism comparable to some of its neighboring Micronesian islands to the east, notably Guam and Pohnpei. -

Republic of Fiji: the State of the World's Forest Genetic Resources
REPUBLIC OF FIJI This country report is prepared as a contribution to the FAO publication, The Report on the State of the World’s Forest Genetic Resources. The content and the structure are in accordance with the recommendations and guidelines given by FAO in the document Guidelines for Preparation of Country Reports for the State of the World’s Forest Genetic Resources (2010). These guidelines set out recommendations for the objective, scope and structure of the country reports. Countries were requested to consider the current state of knowledge of forest genetic diversity, including: Between and within species diversity List of priority species; their roles and values and importance List of threatened/endangered species Threats, opportunities and challenges for the conservation, use and development of forest genetic resources These reports were submitted to FAO as official government documents. The report is presented on www. fao.org/documents as supportive and contextual information to be used in conjunction with other documentation on world forest genetic resources. The content and the views expressed in this report are the responsibility of the entity submitting the report to FAO. FAO may not be held responsible for the use which may be made of the information contained in this report. STATE OF THE FOREST GENETIC RESOURCES IN FIJI Department of Forests Ministry of Fisheries and Forests for The Republic of Fiji Islands and the Secreatriat of Pacific Communities (SPC) State of the Forest Genetic Resources in Fiji _____________________________________________________________________________________________________________________ Table of Contents Executve Summary ………………………………………………………………………………………………………………………..…….. 5 Introduction ………………………………………………………………………………………………………………………………..…….. 6 Chapter 1: The Current State of the Forest Genetic Resources in Fiji ………………………………………………………………….……. -

Trait Correlates and Functional Significance of Heteranthery In
New Research Phytologist Trait correlates and functional significance of heteranthery in flowering plants Mario Vallejo-Marı´n1, Elizabeth M. Da Silva2, Risa D. Sargent2 and Spencer C. H. Barrett3 1School of Biological and Environmental Sciences, University of Stirling, Stirling FK9 4LA, UK; 2Department of Biology, University of Ottawa, 30 Marie-Curie (160 Gendron), Ottawa, ON K1N 6N5, Canada; 3Department of Ecology and Evolutionary Biology, University of Toronto, 25 Willcocks Street, Toronto, ON M5S 3B2, Canada Summary Author for correspondence: • Flowering plants display extraordinary diversity in the morphology of male sexual Mario Vallejo-Marı´n organs, yet the functional significance of this variation is not well understood. Tel: +44 1786 467822 Here, we conducted a comparative analysis of floral correlates of heteranthery – Email: [email protected] the morphological and functional differentiation of anthers within flowers – among Received: 25 June 2010 angiosperm families to identify traits associated with this condition. Accepted: 15 July 2010 • We performed a phylogenetic analysis of correlated evolution between hete- ranthery and several floral traits commonly reported from heterantherous taxa. In New Phytologist (2010) 188: 418–425 addition, we quantified the effect of phylogenetic uncertainty in the observed pat- doi: 10.1111/j.1469-8137.2010.03430.x terns of correlated evolution by comparing trees in which polytomous branches were randomly resolved. • Heteranthery is reported from 12 angiosperm orders and is phylogenetically Key words: buzz-pollination, division of labour, heteranthery, phylogenetic analysis, associated with the absence of floral nectaries, buzz-pollination and enantiostyly stamen differentiation. (mirror-image flowers). These associations are robust to particularities of the underlying phylogenetic hypothesis. -
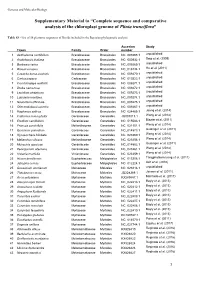
Complete Sequence and Comparative Analysis of the Chloroplast Genome of Plinia Trunciflora”
Genetics and Molecular Biology Supplementary Material to “Complete sequence and comparative analysis of the chloroplast genome of Plinia trunciflora” Table S3 - List of 56 plastome sequences of Rosids included in the Bayesian phylogenetic analysis. Accesion Study Taxon Family Order number 1 Aethionema cordifolium Brassicaceae Brassicales NC_009265.1 unpublished 2 Arabidopsis thaliana Brassicaceae Brassicales NC_000932.1 Sato et al. (1999) 3 Barbarea verna Brassicaceae Brassicales NC_009269.1 unpublished 4 Brassica napus Brassicaceae Brassicales NC_016734.1 Hu et al. (2011) 5 Capsella bursa-pastoris Brassicaceae Brassicales NC_009270.1 unpublished 6 Carica papaya Caricaceae Brassicales NC_010323.1 unpublished 7 Crucihimalaya wallichii Brassicaceae Brassicales NC_009271.1 unpublished 8 Draba nemorosa Brassicaceae Brassicales NC_009272.1 unpublished 9 Lepidium virginicum Brassicaceae Brassicales NC_009273.1 unpublished 10 Lobularia maritima Brassicaceae Brassicales NC_009274.1 unpublished 11 Nasturtium officinale Brassicaceae Brassicales NC_009275.1 unpublished 12 Olimarabidopsis pumila Brassicaceae Brassicales NC_009267.1 unpublished 13 Raphanus sativus Brassicaceae Brassicales NC_024469.1 Jeong et al. (2014) 14 California macrophylla Geraniaceae Geraniales JQ031013.1 Weng et al. (2014) 15 Erodium carvifolium Geraniaceae Geraniales NC_015083.1 Blazier et al. (2011) 16 Francoa sonchifolia Melianthaceae Geraniales NC_021101.1 Weng et al. (2014) 17 Geranium palmatum Geraniaceae Geraniales NC_014573.1 Guisinger et al. (2011) 18 Hypseocharis bilobate Geraniaceae Geraniales NC_023260.1 Weng et al. (2014) 19 Melianthus villosus Melianthaceae Geraniales NC_023256.1 Weng et al. (2014) 20 Monsonia speciose Geraniaceae Geraniales NC_014582.1 Guisinger et al. (2011) 21 Pelargonium alternans Geraniaceae Geraniales NC_023261.1 Weng et al. (2014) 22 Viviania marifolia Vivianiaceae Geraniales NC_023259.1 Weng et al. (2014) 23 Hevea brasiliensis Euphorbiaceae Malpighiales NC_015308.1 Tangphatsornruang et al. -
China's Biodiversity Hotspots Revisited: a Treasure Chest for Plants
A peer-reviewed open-access journal PhytoKeys 130: 1–24 (2019)China’s biodiversity hotspots revisited: A treasure chest for plants 1 doi: 10.3897/phytokeys.130.38417 EDITORIAL http://phytokeys.pensoft.net Launched to accelerate biodiversity research China’s biodiversity hotspots revisited: A treasure chest for plants Jie Cai1, Wen-Bin Yu2,4,5, Ting Zhang1, Hong Wang3, De-Zhu Li1 1 Germplasm Bank of Wild Species, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming, Yunnan 650201, China 2 Center for Integrative Conservation, Xishuangbanna Tropical Botanical Garden, Chinese Academy of Sciences, Mengla, Yunnan 666303, China 3 Key Laboratory for Plant Diversity and Biogeography of East Asia, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming, Yunnan 650201, China 4 Southeast Asia Biodiversity Research Institute, Chinese Academy of Science, Yezin, Nay Pyi Taw 05282, Myanmar 5 Center of Conservation Biology, Core Botanical Gardens, Chinese Academy of Scien- ces, Mengla, Yunnan 666303, China Corresponding author: De-Zhu Li ([email protected]) Received 22 July 2019 | Accepted 12 August 2019 | Published 29 August 2019 Citation: Cai J, Yu W-B, Zhang T, Wang H, Li D-Z (2019) China’s biodiversity hotspots revisited: A treasure chest for plants. In: Cai J, Yu W-B, Zhang T, Li D-Z (Eds) Revealing of the plant diversity in China’s biodiversity hotspots. PhytoKeys 130: 1–24. https://doi.org/10.3897/phytokeys.130.38417 China has been recognised as having exceptionally high plant biodiversity since the mid-19th century, when western plant explorers brought their discoveries to the atten- tion of modern botany (Bretschneider 1898).