Primary Cilia
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Mechanical Forces Induce an Asthma Gene Signature in Healthy Airway Epithelial Cells Ayşe Kılıç1,10, Asher Ameli1,2,10, Jin-Ah Park3,10, Alvin T
www.nature.com/scientificreports OPEN Mechanical forces induce an asthma gene signature in healthy airway epithelial cells Ayşe Kılıç1,10, Asher Ameli1,2,10, Jin-Ah Park3,10, Alvin T. Kho4, Kelan Tantisira1, Marc Santolini 1,5, Feixiong Cheng6,7,8, Jennifer A. Mitchel3, Maureen McGill3, Michael J. O’Sullivan3, Margherita De Marzio1,3, Amitabh Sharma1, Scott H. Randell9, Jefrey M. Drazen3, Jefrey J. Fredberg3 & Scott T. Weiss1,3* Bronchospasm compresses the bronchial epithelium, and this compressive stress has been implicated in asthma pathogenesis. However, the molecular mechanisms by which this compressive stress alters pathways relevant to disease are not well understood. Using air-liquid interface cultures of primary human bronchial epithelial cells derived from non-asthmatic donors and asthmatic donors, we applied a compressive stress and then used a network approach to map resulting changes in the molecular interactome. In cells from non-asthmatic donors, compression by itself was sufcient to induce infammatory, late repair, and fbrotic pathways. Remarkably, this molecular profle of non-asthmatic cells after compression recapitulated the profle of asthmatic cells before compression. Together, these results show that even in the absence of any infammatory stimulus, mechanical compression alone is sufcient to induce an asthma-like molecular signature. Bronchial epithelial cells (BECs) form a physical barrier that protects pulmonary airways from inhaled irritants and invading pathogens1,2. Moreover, environmental stimuli such as allergens, pollutants and viruses can induce constriction of the airways3 and thereby expose the bronchial epithelium to compressive mechanical stress. In BECs, this compressive stress induces structural, biophysical, as well as molecular changes4,5, that interact with nearby mesenchyme6 to cause epithelial layer unjamming1, shedding of soluble factors, production of matrix proteins, and activation matrix modifying enzymes, which then act to coordinate infammatory and remodeling processes4,7–10. -

1714 Gene Comprehensive Cancer Panel Enriched for Clinically Actionable Genes with Additional Biologically Relevant Genes 400-500X Average Coverage on Tumor
xO GENE PANEL 1714 gene comprehensive cancer panel enriched for clinically actionable genes with additional biologically relevant genes 400-500x average coverage on tumor Genes A-C Genes D-F Genes G-I Genes J-L AATK ATAD2B BTG1 CDH7 CREM DACH1 EPHA1 FES G6PC3 HGF IL18RAP JADE1 LMO1 ABCA1 ATF1 BTG2 CDK1 CRHR1 DACH2 EPHA2 FEV G6PD HIF1A IL1R1 JAK1 LMO2 ABCB1 ATM BTG3 CDK10 CRK DAXX EPHA3 FGF1 GAB1 HIF1AN IL1R2 JAK2 LMO7 ABCB11 ATR BTK CDK11A CRKL DBH EPHA4 FGF10 GAB2 HIST1H1E IL1RAP JAK3 LMTK2 ABCB4 ATRX BTRC CDK11B CRLF2 DCC EPHA5 FGF11 GABPA HIST1H3B IL20RA JARID2 LMTK3 ABCC1 AURKA BUB1 CDK12 CRTC1 DCUN1D1 EPHA6 FGF12 GALNT12 HIST1H4E IL20RB JAZF1 LPHN2 ABCC2 AURKB BUB1B CDK13 CRTC2 DCUN1D2 EPHA7 FGF13 GATA1 HLA-A IL21R JMJD1C LPHN3 ABCG1 AURKC BUB3 CDK14 CRTC3 DDB2 EPHA8 FGF14 GATA2 HLA-B IL22RA1 JMJD4 LPP ABCG2 AXIN1 C11orf30 CDK15 CSF1 DDIT3 EPHB1 FGF16 GATA3 HLF IL22RA2 JMJD6 LRP1B ABI1 AXIN2 CACNA1C CDK16 CSF1R DDR1 EPHB2 FGF17 GATA5 HLTF IL23R JMJD7 LRP5 ABL1 AXL CACNA1S CDK17 CSF2RA DDR2 EPHB3 FGF18 GATA6 HMGA1 IL2RA JMJD8 LRP6 ABL2 B2M CACNB2 CDK18 CSF2RB DDX3X EPHB4 FGF19 GDNF HMGA2 IL2RB JUN LRRK2 ACE BABAM1 CADM2 CDK19 CSF3R DDX5 EPHB6 FGF2 GFI1 HMGCR IL2RG JUNB LSM1 ACSL6 BACH1 CALR CDK2 CSK DDX6 EPOR FGF20 GFI1B HNF1A IL3 JUND LTK ACTA2 BACH2 CAMTA1 CDK20 CSNK1D DEK ERBB2 FGF21 GFRA4 HNF1B IL3RA JUP LYL1 ACTC1 BAG4 CAPRIN2 CDK3 CSNK1E DHFR ERBB3 FGF22 GGCX HNRNPA3 IL4R KAT2A LYN ACVR1 BAI3 CARD10 CDK4 CTCF DHH ERBB4 FGF23 GHR HOXA10 IL5RA KAT2B LZTR1 ACVR1B BAP1 CARD11 CDK5 CTCFL DIAPH1 ERCC1 FGF3 GID4 HOXA11 IL6R KAT5 ACVR2A -

Investigations Into Neuronal Cilia Utilizing Mouse Models
INVESTIGATIONS INTO NEURONAL CILIA UTILIZING MOUSE MODELS OF BARDET-BIEDL SYNDROME Dissertation Presented In Partial Fulfillment of the Requirements for the Degree Doctor of Philosophy in the Graduate School of the Ohio State University By Nicolas F. Berbari, BS ***** The Ohio State University 2008 Dissertation Committee: Approved by: Kirk Mykytyn, PhD, Adviser Virginia Sanders, PhD __________________________________________ Georgia Bishop, PhD Adviser Michael Robinson, PhD Integrated Biomedical Sciences Graduate Program ABSTRACT Cilia are hair-like microtubule based cellular appendages that extend 5-30 microns from the surface of most vertebrate cells. Since their initial discovery over a hundred years ago, cilia have been of interest to microbiologists and others studying the dynamics and physiological relevance of their motility. The more recent realization that immotile or primary cilia dysfunction is the basis of several human genetic disorders and diseases has brought the efforts of the biomedical research establishment to bear on this long overlooked and underappreciated organelle. Several human genetic disorders caused by cilia defects have been identified, and include Bardet-Biedl syndrome, Joubert syndrome, Meckel-Gruber syndrome, Alstrom syndrome and orofaciodigital syndrome. One theme of these disorders is their multitude of clinical features such as blindness, cystic kidneys, cognitive deficits and obesity. The fact that many of these cilia disorders present with several features may be due to the ubiquitous nature of the primary cilium and their unrecognized roles in most tissues and cell types. The lack of known function for most primary cilia is no more apparent than in the central nervous system. While it has been known for some time that neurons throughout the brain have primary cilia, their functions remain unknown. -
Type of the Paper (Article
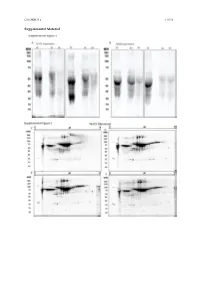
Cells 2020, 9, x 1 of 19 Supplemental Material Cells 2020, 9, x 2 of 19 Figure S1. Secretome enrichment: protocol optimization. 1D SDS-PAGE documentation of washing steps: Culture medium was substituted with FCS-free medium, which was changed every 2 h. The supernatants were then collected, and the proteins isolated and separated in 1D SDS-PAGE ((A) TK173 and (B) TK188). Proteins were stained with Flamingo fluorescent gel stain. Two-dimensional pattern of the proteins isolated from supernatant of TK173, (C) 2 h, (D) 4 h, (E) 6 h, and (F) 8 h after changing to FCS-free medium. (G) Cell secretome collected 24 h after elimination of the contaminating FCS-proteins with different washing steps. Proteins were stained with Flamingo fluorescent gel stain. Cells 2020, 9, x 3 of 19 Figure S2. 2-DE reference maps of secretomes; 150 μg proteins were loaded on an 11 cm IPG strip with a linear pH gradient PI 5–8 for IEF; 12% SDS-polyacrylamide gels were used for the second dimension. Proteins were stained with Flamingo fluorescent gel stain. Identified spots were assigned a number corresponding to that in their table. 2-DE maps from secretome of (A) TK173 control and (B) TGFβ1- treated ones. The 2-DE patterns revealed an alteration of secretome in stimulated TK173. Secretome patterns from TK173 treated with (C) ANG II and (D) PDGF. Cells 2020, 9, x 4 of 19 A Figure S3. Classification of the differentially expressed proteins upon ANG II, TGFβ1, or PDGF treatment in TK173. (A) Bar charts of the cellular component analyzed by STRAP biological function analysis in which the identified proteins from all treatments in both cell types are involved. -

Figure S1. HAEC ROS Production and ML090 NOX5-Inhibition
Figure S1. HAEC ROS production and ML090 NOX5-inhibition. (a) Extracellular H2O2 production in HAEC treated with ML090 at different concentrations and 24 h after being infected with GFP and NOX5-β adenoviruses (MOI 100). **p< 0.01, and ****p< 0.0001 vs control NOX5-β-infected cells (ML090, 0 nM). Results expressed as mean ± SEM. Fold increase vs GFP-infected cells with 0 nM of ML090. n= 6. (b) NOX5-β overexpression and DHE oxidation in HAEC. Representative images from three experiments are shown. Intracellular superoxide anion production of HAEC 24 h after infection with GFP and NOX5-β adenoviruses at different MOIs treated or not with ML090 (10 nM). MOI: Multiplicity of infection. Figure S2. Ontology analysis of HAEC infected with NOX5-β. Ontology analysis shows that the response to unfolded protein is the most relevant. Figure S3. UPR mRNA expression in heart of infarcted transgenic mice. n= 12-13. Results expressed as mean ± SEM. Table S1: Altered gene expression due to NOX5-β expression at 12 h (bold, highlighted in yellow). N12hvsG12h N18hvsG18h N24hvsG24h GeneName GeneDescription TranscriptID logFC p-value logFC p-value logFC p-value family with sequence similarity NM_052966 1.45 1.20E-17 2.44 3.27E-19 2.96 6.24E-21 FAM129A 129. member A DnaJ (Hsp40) homolog. NM_001130182 2.19 9.83E-20 2.94 2.90E-19 3.01 1.68E-19 DNAJA4 subfamily A. member 4 phorbol-12-myristate-13-acetate- NM_021127 0.93 1.84E-12 2.41 1.32E-17 2.69 1.43E-18 PMAIP1 induced protein 1 E2F7 E2F transcription factor 7 NM_203394 0.71 8.35E-11 2.20 2.21E-17 2.48 1.84E-18 DnaJ (Hsp40) homolog. -

Extending Our Understanding of Cardiovascular Risk From
EDITORIALS www.jasn.org multiprotein cilia-destined cargo within the cell. Then through 13. Tyler KM, Fridberg A, Toriello KM, Olson CL, Cieslak JA, Hazlett TL, association of an adaptor molecule containing a cilia-targeting Engman DM: Flagellar membrane localization via association with lipid rafts. J Cell Sci 122: 859–866, 2009 motif, perhaps such as cystin, multiple proteins deliver to the 14. Rohatgi R, Milenkovic L, Scott MP: Patched1 regulates hedgehog cilium. Further complexity in this model arises from the fact signaling at the primary cilium. Science 317: 372–376, 2007 that not all proteins constitutively localize to cilia but require an external stimulus that promotes cilia entry. A good example of this is Hedgehog signaling. It was recently shown upon bind- See related article, “Cystin Localizes to Primary Cilia via Membrane Microdo- ing of Hedgehog to its receptor Patched, the protein Smooth- mains and a Targeting Motif,” on pages 2570–2580. ened is targeted to the cilium14; therefore, multiple targeting motifs and adaptors likely regulate ciliary entry of proteins either in a constitutive manner or in response to a specific cue. Understanding these targeting signals will be key as we con- It’s about Time: Extending our tinue to unravel the mysteries of the cilia. Understanding of Cardiovascular Risk from DISCLOSURES Chronic Kidney Disease This work was supported in part by National Institutes of Health (NIH DK069605) and Polycystic Kidney Disease Foundation (162G08a) grants. Ann Young and Amit X. Garg Division of Nephrology and Department of Epidemiology and Bio- statistics, University of Western Ontario, London, Ontario, Canada REFERENCES J Am Soc Nephrol 20: 2486–2487, 2009. -

Polycystin-2 Takes Different Routes to the Somatic and Ciliary Plasma Membrane
JCB: Article Polycystin-2 takes different routes to the somatic and ciliary plasma membrane Helen Hoffmeister,1 Karin Babinger,1 Sonja Gürster,1 Anna Cedzich,1,2 Christine Meese,1 Karin Schadendorf,3 Larissa Osten,1 Uwe de Vries,1 Anne Rascle,1 and Ralph Witzgall1 1Institute for Molecular and Cellular Anatomy, University of Regensburg, 93053 Regensburg, Germany 2Medical Research Center, Klinikum Mannheim, University of Heidelberg, 68167 Mannheim, Germany 3Center for Electron Microscopy, University of Regensburg, 93053 Regensburg, Germany olycystin-2 (also called TRPP2), an integral mem- a COPII-dependent fashion at the endoplasmic reticulum, brane protein mutated in patients with cystic kidney that polycystin-2 reaches the cis side of the Golgi appara- P disease, is located in the primary cilium where it is tus in either case, but that the trafficking to the somatic thought to transmit mechanical stimuli into the cell interior. plasma membrane goes through the Golgi apparatus After studying a series of polycystin-2 deletion mutants we whereas transport vesicles to the cilium leave the Golgi identified two amino acids in loop 4 that were essential for apparatus at the cis compartment. Such an interpretation the trafficking of polycystin-2 to the somatic (nonciliary) is supported by the finding that mycophenolic acid treat- plasma membrane. However, polycystin-2 mutant proteins ment resulted in the colocalization of polycystin-2 with in which these two residues were replaced by alanine GM130, a marker of the cis-Golgi apparatus. Remark- were still sorted into the cilium, thus indicating that the ably, we also observed that wild-type Smoothened, an trafficking routes to the somatic and ciliary plasma mem- integral membrane protein involved in hedgehog signaling brane compartments are distinct. -
The Transition Zone: an Essential Functional Compartment of Cilia Szymanska and Johnson
The transition zone: an essential functional compartment of cilia Szymanska and Johnson Szymanska and Johnson Cilia 2012, 1:10 http://www.ciliajournal.com/content/1/1/10 Szymanska and Johnson Cilia 2012, 1:10 http://www.ciliajournal.com/content/1/1/10 REVIEW Open Access The transition zone: an essential functional compartment of cilia Katarzyna Szymanska and Colin A Johnson* Abstract Recent studies of the primary cilium have begun to provide further insights into ciliary ultrastructure, with an emerging picture of complex compartmentalization and molecular components that combine in functional modules. Many proteins that are mutated in ciliopathies are localized to the transition zone, a compartment of the proximal region of the cilium. The loss of these components can disrupt ciliary functions such as the control of protein entry and exit from the cilium, the possible trafficking of essential ciliary components, and the regulation of signaling cascades and control of the cell cycle. The discovery of functional modules within the primary cilium may help in understanding the variable phenotypes and pleiotropy in ciliopathies. Keywords: Cilia, Transition zone, IFT, Ciliopathies Review crucial role in left-right patterning. They are located at Cilia are microtubule-based, hair-like organelles that the embryonic node, and although they have a “9+0” occur on the apical surface of most mammalian cells in microtubule pattern they mediate a leftward flow at the G0/G1 of the cell cycle [1], with the exception of bone node in a “whirlpool”-likemannerduetotheretention marrow-derived cells [2]. Defects in cilia structure or of dynein arms between microtubules. The leftward function are the cause of a suite of congenital conditions flow is thought to transport morphogens that are essen- known as ciliopathies, which now include polycystic kid- tial for the first step of symmetry breaking in the devel- ney disease, nephronophthisis, Senior-Løken syndrome, opmental of the mammalian body plan [2]. -

The Ciliopathy Gene Nphp-2 Functions in Multiple Gene Networks and Regulates
THE CILIOPATHY GENE NPHP-2 FUNCTIONS IN MULTIPLE GENE NETWORKS AND REGULATES CILIOGENESIS IN C. ELEGANS By SIMON ROBERT FREDERICK WARBURTON-PITT A dissertation submitted to the Graduate School – New Brunswick and The Graduate School of Biomedical Sciences Rutgers, The State University of New Jersey In partial fulfillment of the requirements For the degree of Doctor of Philosophy Graduate Program in Molecular Genetics and Microbiology Written under the direction of Maureen M. Barr, PhD And approved by New Brunswick, New Jersey January, 2015 © 2015 Simon Warburton-Pitt ALL RIGHTS RESERVED ABSTRACT OF THE DISSERTATION The ciliopathy gene nphp-2 functions in multiple gene networks and regulates ciliogenesis in C. elegans By SIMON ROBERT FREDERICK WARBURTON-PITT Dissertation Director: Maureen Barr Cilia are hair-like organelles that function as cellular antennae. Cilia are conserved across eukaryotes, and play a vital role in many biological processes including signal transduction, signal cascades, cell-cell signaling, cell orientation, cell-cell adhesion, motility, interorganismal communication, building extracellular matrix, and inducing fluid flow. In humans, cilia are present in a majority of tissue types, and cilia dysfunction can lead to a range of syndromic ciliopathies, including nephronopthisis (NPHP) and Meckel Syndrome (MKS). Cilia have a microtubule backbone, the axoneme, and are composed of multiple subcompartments, each with a specific function and composition: the transition zone (TZ) anchoring to the axoneme to the membrane, the doublet region extending from the TZ, and in some cilia types, the singlet region extending from the doublet region. The nematode C. elegans is a well-established model of cilia biology, and possesses cilia at the distal end of sensory dendrites. -

Cystin, Cilia, and Cysts: Unraveling Trafficking Determinants
EDITORIALS www.jasn.org UP FRONT MATTERS Cystin, Cilia, and Cysts: cilia, and photoreceptor outer segments, respectively.5–7 In photoreceptors, this motif binds the small G-protein, Arf4, Unraveling Trafficking and in conjunction with Rab11, FIP3, and ASAP1 promotes budding of ciliary-destined cargo from the transgolgi net- Determinants work.7 8 † In this issue of JASN, Tao et al. identify a novel ciliary Toby W. Hurd* and Ben Margolis trafficking determinant in the cystoprotein cystin that fur- Departments of *Pediatrics and †Internal Medicine, University of Michigan, Ann Arbor, Michigan thers our understanding of how proteins are selectively tar- geted to the cilium. This group previously identified cystin as J Am Soc Nephrol ●●: –, 2009. the gene disrupted in the cpk (congenital polycystic kidney) doi: 10.1681/ASN.2009090996 mouse model of autosomal recessive PKD.9 Of particular in- terest is the previous observation that the cystin protein lo- The past 10 yr has seen an exponential increase in research calizes to renal cilia3; however, little was known about the into a previously often ignored organelle, the primary cilium. function of this protein or the mechanism by which it traffics This is largely as a result of the discovery that ciliary dysfunc- to the cilium. tion underlies many human inheritable disorders, termed cil- Sequence and domain analyses of cystin yield no sequence iopathies.1 These include polycystic kidney disease (PKD), homology to other proteins, although it is predicted to be nephronophthisis, and retinal degeneration. Cilia are micro- myristoylated at the amino-terminus. Tao et al.8 test this hy- tubule-based organelles whose core components, the in- pothesis by in vivo labeling and indeed prove this is the case. -

Origin, Characterization and Roles of Matrix Vesicles in Physiological and Pathological Mineralization
Origin, characterization and roles of matrix vesicles in physiological and pathological mineralization. Cyril Thouverey To cite this version: Cyril Thouverey. Origin, characterization and roles of matrix vesicles in physiological and pathological mineralization.. Life Sciences [q-bio]. Université Claude Bernard - Lyon I, 2008. English. tel- 00304214 HAL Id: tel-00304214 https://tel.archives-ouvertes.fr/tel-00304214 Submitted on 22 Jul 2008 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. N° d’ordre: 95-2008 Année 2008 THESE Présentée devant Nencki Institute of Experimental Biology, Polish Academy of Sciences Université Claude Bernard Lyon 1, Pour l’obtention Du DIPLOME DE DOCTORAT (Arrêté du 7 août 2006 et arrêté du 6 janvier 2005) Présentée et soutenue publiquement le 20 juin 2008 Par Cyril Thouverey Origin, characterization and roles of matrix vesicles in physiological and pathological mineralization Directeurs de thèse: Pr René Buchet et Pr Sławomir Pikuła JURY: Mme Françoise Bleicher Mme Joanna Bandorowicz-Pikuła Mme Martime Cohen-Solal Mr René Buchet Mr Sławomir Pikuła Mr Aleksander Sikorski UNIVERSITE CLAUDE BERNARD - LYON I Président de l’Université M. le Professeur L. COLLET Vice-Président du Conseil Scientifique M. -

Expression of Primary Cilia-Related Genes in Developing Mouse Gonads RAFAL P
Int. J. Dev. Biol. 63: 615-621 (2019) https://doi.org/10.1387/ijdb.190049rp www.intjdevbiol.com Expression of primary cilia-related genes in developing mouse gonads RAFAL P. PIPREK*,1, DAGMARA PODKOWA1, MALGORZATA KLOC2,3,4 and JACEK Z. KUBIAK5,6 1Department of Comparative Anatomy, Institute of Zoology and Biomedical Research, Jagiellonian University, Krakow, Poland, 2The Houston Methodist Research Institute, Houston, TX, USA, 3Department of Surgery, The Houston Methodist Hospital, Houston TX, USA, 4University of Texas, MD Anderson Cancer Center, Houston TX, USA, 5Univ Rennes, CNRS, Institute of Genetics and Development of Rennes, UMR 6290, Cell Cycle Group, Faculty of Medicine, Rennes, France and 6Laboratory of Regenerative Medicine and Cell Biology, Military Institute of Hygiene and Epidemiology (WIHE), Warsaw, Poland ABSTRACT Mechanisms governing differentiation of the bipotential gonad into the testes or ovaries are complex and still vague. The primary cilium is an organelle involved in cell signaling, which controls the development of many organs, but the role of primary cilium in the sex determination and sexual differentiation of gonads is completely unknown. Here we studied the expression of genes involved in primary cilium formation and functioning in fetal mouse gonads, before, during and after sexual differentiation. We studied the expression of 175 primary cilia-related genes using microarray technique. 144 of these genes were ubiquitously expressed in all studied cell types with no significant differences in expression level. Such a high level of expression of primary cilia-related genes in developing mouse gonads suggests that the primary cilia and/or primary cilia-related genes are important for the development of both somatic and germline component of the gonads.