Functional Study of Mir-27A in Human Hepatic Stellate Cells by Proteomic Analysis: Comprehensive View and a Role in Myogenic Tans-Differentiation
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Analysis of Trans Esnps Infers Regulatory Network Architecture
Analysis of trans eSNPs infers regulatory network architecture Anat Kreimer Submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy in the Graduate School of Arts and Sciences COLUMBIA UNIVERSITY 2014 © 2014 Anat Kreimer All rights reserved ABSTRACT Analysis of trans eSNPs infers regulatory network architecture Anat Kreimer eSNPs are genetic variants associated with transcript expression levels. The characteristics of such variants highlight their importance and present a unique opportunity for studying gene regulation. eSNPs affect most genes and their cell type specificity can shed light on different processes that are activated in each cell. They can identify functional variants by connecting SNPs that are implicated in disease to a molecular mechanism. Examining eSNPs that are associated with distal genes can provide insights regarding the inference of regulatory networks but also presents challenges due to the high statistical burden of multiple testing. Such association studies allow: simultaneous investigation of many gene expression phenotypes without assuming any prior knowledge and identification of unknown regulators of gene expression while uncovering directionality. This thesis will focus on such distal eSNPs to map regulatory interactions between different loci and expose the architecture of the regulatory network defined by such interactions. We develop novel computational approaches and apply them to genetics-genomics data in human. We go beyond pairwise interactions to define network motifs, including regulatory modules and bi-fan structures, showing them to be prevalent in real data and exposing distinct attributes of such arrangements. We project eSNP associations onto a protein-protein interaction network to expose topological properties of eSNPs and their targets and highlight different modes of distal regulation. -

Mechanical Forces Induce an Asthma Gene Signature in Healthy Airway Epithelial Cells Ayşe Kılıç1,10, Asher Ameli1,2,10, Jin-Ah Park3,10, Alvin T
www.nature.com/scientificreports OPEN Mechanical forces induce an asthma gene signature in healthy airway epithelial cells Ayşe Kılıç1,10, Asher Ameli1,2,10, Jin-Ah Park3,10, Alvin T. Kho4, Kelan Tantisira1, Marc Santolini 1,5, Feixiong Cheng6,7,8, Jennifer A. Mitchel3, Maureen McGill3, Michael J. O’Sullivan3, Margherita De Marzio1,3, Amitabh Sharma1, Scott H. Randell9, Jefrey M. Drazen3, Jefrey J. Fredberg3 & Scott T. Weiss1,3* Bronchospasm compresses the bronchial epithelium, and this compressive stress has been implicated in asthma pathogenesis. However, the molecular mechanisms by which this compressive stress alters pathways relevant to disease are not well understood. Using air-liquid interface cultures of primary human bronchial epithelial cells derived from non-asthmatic donors and asthmatic donors, we applied a compressive stress and then used a network approach to map resulting changes in the molecular interactome. In cells from non-asthmatic donors, compression by itself was sufcient to induce infammatory, late repair, and fbrotic pathways. Remarkably, this molecular profle of non-asthmatic cells after compression recapitulated the profle of asthmatic cells before compression. Together, these results show that even in the absence of any infammatory stimulus, mechanical compression alone is sufcient to induce an asthma-like molecular signature. Bronchial epithelial cells (BECs) form a physical barrier that protects pulmonary airways from inhaled irritants and invading pathogens1,2. Moreover, environmental stimuli such as allergens, pollutants and viruses can induce constriction of the airways3 and thereby expose the bronchial epithelium to compressive mechanical stress. In BECs, this compressive stress induces structural, biophysical, as well as molecular changes4,5, that interact with nearby mesenchyme6 to cause epithelial layer unjamming1, shedding of soluble factors, production of matrix proteins, and activation matrix modifying enzymes, which then act to coordinate infammatory and remodeling processes4,7–10. -

NICU Gene List Generator.Xlsx
Neonatal Crisis Sequencing Panel Gene List Genes: A2ML1 - B3GLCT A2ML1 ADAMTS9 ALG1 ARHGEF15 AAAS ADAMTSL2 ALG11 ARHGEF9 AARS1 ADAR ALG12 ARID1A AARS2 ADARB1 ALG13 ARID1B ABAT ADCY6 ALG14 ARID2 ABCA12 ADD3 ALG2 ARL13B ABCA3 ADGRG1 ALG3 ARL6 ABCA4 ADGRV1 ALG6 ARMC9 ABCB11 ADK ALG8 ARPC1B ABCB4 ADNP ALG9 ARSA ABCC6 ADPRS ALK ARSL ABCC8 ADSL ALMS1 ARX ABCC9 AEBP1 ALOX12B ASAH1 ABCD1 AFF3 ALOXE3 ASCC1 ABCD3 AFF4 ALPK3 ASH1L ABCD4 AFG3L2 ALPL ASL ABHD5 AGA ALS2 ASNS ACAD8 AGK ALX3 ASPA ACAD9 AGL ALX4 ASPM ACADM AGPS AMELX ASS1 ACADS AGRN AMER1 ASXL1 ACADSB AGT AMH ASXL3 ACADVL AGTPBP1 AMHR2 ATAD1 ACAN AGTR1 AMN ATL1 ACAT1 AGXT AMPD2 ATM ACE AHCY AMT ATP1A1 ACO2 AHDC1 ANK1 ATP1A2 ACOX1 AHI1 ANK2 ATP1A3 ACP5 AIFM1 ANKH ATP2A1 ACSF3 AIMP1 ANKLE2 ATP5F1A ACTA1 AIMP2 ANKRD11 ATP5F1D ACTA2 AIRE ANKRD26 ATP5F1E ACTB AKAP9 ANTXR2 ATP6V0A2 ACTC1 AKR1D1 AP1S2 ATP6V1B1 ACTG1 AKT2 AP2S1 ATP7A ACTG2 AKT3 AP3B1 ATP8A2 ACTL6B ALAS2 AP3B2 ATP8B1 ACTN1 ALB AP4B1 ATPAF2 ACTN2 ALDH18A1 AP4M1 ATR ACTN4 ALDH1A3 AP4S1 ATRX ACVR1 ALDH3A2 APC AUH ACVRL1 ALDH4A1 APTX AVPR2 ACY1 ALDH5A1 AR B3GALNT2 ADA ALDH6A1 ARFGEF2 B3GALT6 ADAMTS13 ALDH7A1 ARG1 B3GAT3 ADAMTS2 ALDOB ARHGAP31 B3GLCT Updated: 03/15/2021; v.3.6 1 Neonatal Crisis Sequencing Panel Gene List Genes: B4GALT1 - COL11A2 B4GALT1 C1QBP CD3G CHKB B4GALT7 C3 CD40LG CHMP1A B4GAT1 CA2 CD59 CHRNA1 B9D1 CA5A CD70 CHRNB1 B9D2 CACNA1A CD96 CHRND BAAT CACNA1C CDAN1 CHRNE BBIP1 CACNA1D CDC42 CHRNG BBS1 CACNA1E CDH1 CHST14 BBS10 CACNA1F CDH2 CHST3 BBS12 CACNA1G CDK10 CHUK BBS2 CACNA2D2 CDK13 CILK1 BBS4 CACNB2 CDK5RAP2 -

Molecular Effects of Isoflavone Supplementation Human Intervention Studies and Quantitative Models for Risk Assessment
Molecular effects of isoflavone supplementation Human intervention studies and quantitative models for risk assessment Vera van der Velpen Thesis committee Promotors Prof. Dr Pieter van ‘t Veer Professor of Nutritional Epidemiology Wageningen University Prof. Dr Evert G. Schouten Emeritus Professor of Epidemiology and Prevention Wageningen University Co-promotors Dr Anouk Geelen Assistant professor, Division of Human Nutrition Wageningen University Dr Lydia A. Afman Assistant professor, Division of Human Nutrition Wageningen University Other members Prof. Dr Jaap Keijer, Wageningen University Dr Hubert P.J.M. Noteborn, Netherlands Food en Consumer Product Safety Authority Prof. Dr Yvonne T. van der Schouw, UMC Utrecht Dr Wendy L. Hall, King’s College London This research was conducted under the auspices of the Graduate School VLAG (Advanced studies in Food Technology, Agrobiotechnology, Nutrition and Health Sciences). Molecular effects of isoflavone supplementation Human intervention studies and quantitative models for risk assessment Vera van der Velpen Thesis submitted in fulfilment of the requirements for the degree of doctor at Wageningen University by the authority of the Rector Magnificus Prof. Dr M.J. Kropff, in the presence of the Thesis Committee appointed by the Academic Board to be defended in public on Friday 20 June 2014 at 13.30 p.m. in the Aula. Vera van der Velpen Molecular effects of isoflavone supplementation: Human intervention studies and quantitative models for risk assessment 154 pages PhD thesis, Wageningen University, Wageningen, NL (2014) With references, with summaries in Dutch and English ISBN: 978-94-6173-952-0 ABSTRact Background: Risk assessment can potentially be improved by closely linked experiments in the disciplines of epidemiology and toxicology. -

Targeting Oncogenic Notch Signaling with SERCA Inhibitors Luca Pagliaro, Matteo Marchesini and Giovanni Roti*
Pagliaro et al. J Hematol Oncol (2021) 14:8 https://doi.org/10.1186/s13045-020-01015-9 REVIEW Open Access Targeting oncogenic Notch signaling with SERCA inhibitors Luca Pagliaro, Matteo Marchesini and Giovanni Roti* Abstract P-type ATPase inhibitors are among the most successful and widely prescribed therapeutics in modern pharmacol- ogy. Clinical transition has been safely achieved for H+/K+ ATPase inhibitors such as omeprazole and Na+/K+-ATPase 2 inhibitors like digoxin. However, this is more challenging for Ca +-ATPase modulators due to the physiological role of 2 2 Ca + in cardiac dynamics. Over the past two decades, sarco-endoplasmic reticulum Ca +-ATPase (SERCA) modula- 2 tors have been studied as potential chemotherapy agents because of their Ca +-mediated pan-cancer lethal efects. Instead, recent evidence suggests that SERCA inhibition suppresses oncogenic Notch1 signaling emerging as an alternative to γ-secretase modulators that showed limited clinical activity due to severe side efects. In this review, we focus on how SERCA inhibitors alter Notch1 signaling and show that Notch on-target-mediated antileukemia proper- 2 ties of these molecules can be achieved without causing overt Ca + cellular overload. Keywords: SERCA , T cell acute lymphoblastic leukemia, Thapsigargin, Notch signaling, NOTCH1, CAD204520, T-ALL Background metalloprotease (ADAM-10 or TACE/ADAM-17). Te NOTCH receptors are transmembrane cell-surface pro- resulting short-lived protein fragments are substrates teins that control cell to cell communication, embryo- -

Supplementary Table S4. FGA Co-Expressed Gene List in LUAD
Supplementary Table S4. FGA co-expressed gene list in LUAD tumors Symbol R Locus Description FGG 0.919 4q28 fibrinogen gamma chain FGL1 0.635 8p22 fibrinogen-like 1 SLC7A2 0.536 8p22 solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 DUSP4 0.521 8p12-p11 dual specificity phosphatase 4 HAL 0.51 12q22-q24.1histidine ammonia-lyase PDE4D 0.499 5q12 phosphodiesterase 4D, cAMP-specific FURIN 0.497 15q26.1 furin (paired basic amino acid cleaving enzyme) CPS1 0.49 2q35 carbamoyl-phosphate synthase 1, mitochondrial TESC 0.478 12q24.22 tescalcin INHA 0.465 2q35 inhibin, alpha S100P 0.461 4p16 S100 calcium binding protein P VPS37A 0.447 8p22 vacuolar protein sorting 37 homolog A (S. cerevisiae) SLC16A14 0.447 2q36.3 solute carrier family 16, member 14 PPARGC1A 0.443 4p15.1 peroxisome proliferator-activated receptor gamma, coactivator 1 alpha SIK1 0.435 21q22.3 salt-inducible kinase 1 IRS2 0.434 13q34 insulin receptor substrate 2 RND1 0.433 12q12 Rho family GTPase 1 HGD 0.433 3q13.33 homogentisate 1,2-dioxygenase PTP4A1 0.432 6q12 protein tyrosine phosphatase type IVA, member 1 C8orf4 0.428 8p11.2 chromosome 8 open reading frame 4 DDC 0.427 7p12.2 dopa decarboxylase (aromatic L-amino acid decarboxylase) TACC2 0.427 10q26 transforming, acidic coiled-coil containing protein 2 MUC13 0.422 3q21.2 mucin 13, cell surface associated C5 0.412 9q33-q34 complement component 5 NR4A2 0.412 2q22-q23 nuclear receptor subfamily 4, group A, member 2 EYS 0.411 6q12 eyes shut homolog (Drosophila) GPX2 0.406 14q24.1 glutathione peroxidase -

Aneuploidy: Using Genetic Instability to Preserve a Haploid Genome?
Health Science Campus FINAL APPROVAL OF DISSERTATION Doctor of Philosophy in Biomedical Science (Cancer Biology) Aneuploidy: Using genetic instability to preserve a haploid genome? Submitted by: Ramona Ramdath In partial fulfillment of the requirements for the degree of Doctor of Philosophy in Biomedical Science Examination Committee Signature/Date Major Advisor: David Allison, M.D., Ph.D. Academic James Trempe, Ph.D. Advisory Committee: David Giovanucci, Ph.D. Randall Ruch, Ph.D. Ronald Mellgren, Ph.D. Senior Associate Dean College of Graduate Studies Michael S. Bisesi, Ph.D. Date of Defense: April 10, 2009 Aneuploidy: Using genetic instability to preserve a haploid genome? Ramona Ramdath University of Toledo, Health Science Campus 2009 Dedication I dedicate this dissertation to my grandfather who died of lung cancer two years ago, but who always instilled in us the value and importance of education. And to my mom and sister, both of whom have been pillars of support and stimulating conversations. To my sister, Rehanna, especially- I hope this inspires you to achieve all that you want to in life, academically and otherwise. ii Acknowledgements As we go through these academic journeys, there are so many along the way that make an impact not only on our work, but on our lives as well, and I would like to say a heartfelt thank you to all of those people: My Committee members- Dr. James Trempe, Dr. David Giovanucchi, Dr. Ronald Mellgren and Dr. Randall Ruch for their guidance, suggestions, support and confidence in me. My major advisor- Dr. David Allison, for his constructive criticism and positive reinforcement. -

1714 Gene Comprehensive Cancer Panel Enriched for Clinically Actionable Genes with Additional Biologically Relevant Genes 400-500X Average Coverage on Tumor
xO GENE PANEL 1714 gene comprehensive cancer panel enriched for clinically actionable genes with additional biologically relevant genes 400-500x average coverage on tumor Genes A-C Genes D-F Genes G-I Genes J-L AATK ATAD2B BTG1 CDH7 CREM DACH1 EPHA1 FES G6PC3 HGF IL18RAP JADE1 LMO1 ABCA1 ATF1 BTG2 CDK1 CRHR1 DACH2 EPHA2 FEV G6PD HIF1A IL1R1 JAK1 LMO2 ABCB1 ATM BTG3 CDK10 CRK DAXX EPHA3 FGF1 GAB1 HIF1AN IL1R2 JAK2 LMO7 ABCB11 ATR BTK CDK11A CRKL DBH EPHA4 FGF10 GAB2 HIST1H1E IL1RAP JAK3 LMTK2 ABCB4 ATRX BTRC CDK11B CRLF2 DCC EPHA5 FGF11 GABPA HIST1H3B IL20RA JARID2 LMTK3 ABCC1 AURKA BUB1 CDK12 CRTC1 DCUN1D1 EPHA6 FGF12 GALNT12 HIST1H4E IL20RB JAZF1 LPHN2 ABCC2 AURKB BUB1B CDK13 CRTC2 DCUN1D2 EPHA7 FGF13 GATA1 HLA-A IL21R JMJD1C LPHN3 ABCG1 AURKC BUB3 CDK14 CRTC3 DDB2 EPHA8 FGF14 GATA2 HLA-B IL22RA1 JMJD4 LPP ABCG2 AXIN1 C11orf30 CDK15 CSF1 DDIT3 EPHB1 FGF16 GATA3 HLF IL22RA2 JMJD6 LRP1B ABI1 AXIN2 CACNA1C CDK16 CSF1R DDR1 EPHB2 FGF17 GATA5 HLTF IL23R JMJD7 LRP5 ABL1 AXL CACNA1S CDK17 CSF2RA DDR2 EPHB3 FGF18 GATA6 HMGA1 IL2RA JMJD8 LRP6 ABL2 B2M CACNB2 CDK18 CSF2RB DDX3X EPHB4 FGF19 GDNF HMGA2 IL2RB JUN LRRK2 ACE BABAM1 CADM2 CDK19 CSF3R DDX5 EPHB6 FGF2 GFI1 HMGCR IL2RG JUNB LSM1 ACSL6 BACH1 CALR CDK2 CSK DDX6 EPOR FGF20 GFI1B HNF1A IL3 JUND LTK ACTA2 BACH2 CAMTA1 CDK20 CSNK1D DEK ERBB2 FGF21 GFRA4 HNF1B IL3RA JUP LYL1 ACTC1 BAG4 CAPRIN2 CDK3 CSNK1E DHFR ERBB3 FGF22 GGCX HNRNPA3 IL4R KAT2A LYN ACVR1 BAI3 CARD10 CDK4 CTCF DHH ERBB4 FGF23 GHR HOXA10 IL5RA KAT2B LZTR1 ACVR1B BAP1 CARD11 CDK5 CTCFL DIAPH1 ERCC1 FGF3 GID4 HOXA11 IL6R KAT5 ACVR2A -
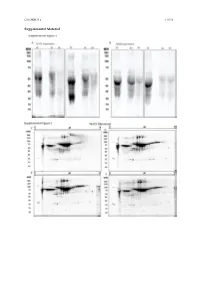
Type of the Paper (Article
Cells 2020, 9, x 1 of 19 Supplemental Material Cells 2020, 9, x 2 of 19 Figure S1. Secretome enrichment: protocol optimization. 1D SDS-PAGE documentation of washing steps: Culture medium was substituted with FCS-free medium, which was changed every 2 h. The supernatants were then collected, and the proteins isolated and separated in 1D SDS-PAGE ((A) TK173 and (B) TK188). Proteins were stained with Flamingo fluorescent gel stain. Two-dimensional pattern of the proteins isolated from supernatant of TK173, (C) 2 h, (D) 4 h, (E) 6 h, and (F) 8 h after changing to FCS-free medium. (G) Cell secretome collected 24 h after elimination of the contaminating FCS-proteins with different washing steps. Proteins were stained with Flamingo fluorescent gel stain. Cells 2020, 9, x 3 of 19 Figure S2. 2-DE reference maps of secretomes; 150 μg proteins were loaded on an 11 cm IPG strip with a linear pH gradient PI 5–8 for IEF; 12% SDS-polyacrylamide gels were used for the second dimension. Proteins were stained with Flamingo fluorescent gel stain. Identified spots were assigned a number corresponding to that in their table. 2-DE maps from secretome of (A) TK173 control and (B) TGFβ1- treated ones. The 2-DE patterns revealed an alteration of secretome in stimulated TK173. Secretome patterns from TK173 treated with (C) ANG II and (D) PDGF. Cells 2020, 9, x 4 of 19 A Figure S3. Classification of the differentially expressed proteins upon ANG II, TGFβ1, or PDGF treatment in TK173. (A) Bar charts of the cellular component analyzed by STRAP biological function analysis in which the identified proteins from all treatments in both cell types are involved. -

Transcriptomic Profiling of Ca Transport Systems During
cells Article Transcriptomic Profiling of Ca2+ Transport Systems during the Formation of the Cerebral Cortex in Mice Alexandre Bouron Genetics and Chemogenomics Lab, Université Grenoble Alpes, CNRS, CEA, INSERM, Bâtiment C3, 17 rue des Martyrs, 38054 Grenoble, France; [email protected] Received: 29 June 2020; Accepted: 24 July 2020; Published: 29 July 2020 Abstract: Cytosolic calcium (Ca2+) transients control key neural processes, including neurogenesis, migration, the polarization and growth of neurons, and the establishment and maintenance of synaptic connections. They are thus involved in the development and formation of the neural system. In this study, a publicly available whole transcriptome sequencing (RNA-Seq) dataset was used to examine the expression of genes coding for putative plasma membrane and organellar Ca2+-transporting proteins (channels, pumps, exchangers, and transporters) during the formation of the cerebral cortex in mice. Four ages were considered: embryonic days 11 (E11), 13 (E13), and 17 (E17), and post-natal day 1 (PN1). This transcriptomic profiling was also combined with live-cell Ca2+ imaging recordings to assess the presence of functional Ca2+ transport systems in E13 neurons. The most important Ca2+ routes of the cortical wall at the onset of corticogenesis (E11–E13) were TACAN, GluK5, nAChR β2, Cav3.1, Orai3, transient receptor potential cation channel subfamily M member 7 (TRPM7) non-mitochondrial Na+/Ca2+ exchanger 2 (NCX2), and the connexins CX43/CX45/CX37. Hence, transient receptor potential cation channel mucolipin subfamily member 1 (TRPML1), transmembrane protein 165 (TMEM165), and Ca2+ “leak” channels are prominent intracellular Ca2+ pathways. The Ca2+ pumps sarco/endoplasmic reticulum Ca2+ ATPase 2 (SERCA2) and plasma membrane Ca2+ ATPase 1 (PMCA1) control the resting basal Ca2+ levels. -

The H3K9 Methylation Writer SETDB1 and Its Reader MPP8 Cooperate to Silence Satellite DNA Repeats in Mouse Embryonic Stem Cells
G C A T T A C G G C A T genes Article The H3K9 Methylation Writer SETDB1 and Its Reader MPP8 Cooperate to Silence Satellite DNA Repeats in Mouse Embryonic Stem Cells 1,2,3,4 1 1, 1 Paola Cruz-Tapias , Philippe Robin , Julien Pontis y, Laurence Del Maestro and Slimane Ait-Si-Ali 1,* 1 Epigenetics and Cell Fate (EDC), Centre National de la Recherche Scientifique (CNRS), Université de Paris, Université Paris Diderot, F-75013 Paris, France; [email protected] (P.C.-T.); [email protected] (P.R.); julien.pontis@epfl.ch (J.P.); [email protected] (L.D.M.) 2 Faculty of Natural Sciences and Mathematics, Universidad del Rosario, Bogotá 111221, Colombia 3 School of Medicine and Health Sciences, Universidad del Rosario, Bogotá 111221, Colombia 4 Doctoral Program in Biomedical and Biological Sciences, Universidad del Rosario, Bogotá 111221, Colombia * Correspondence: [email protected]; Tel.: +33-(0)1-5727-8919 Current: Ecole Polytechnique Fédérale de Lausanne (EPFL), SV LVG Station 19, 1015 Lausanne, Switzerland. y Received: 25 August 2019; Accepted: 24 September 2019; Published: 25 September 2019 Abstract: SETDB1 (SET Domain Bifurcated histone lysine methyltransferase 1) is a key lysine methyltransferase (KMT) required in embryonic stem cells (ESCs), where it silences transposable elements and DNA repeats via histone H3 lysine 9 tri-methylation (H3K9me3), independently of DNA methylation. The H3K9 methylation reader M-Phase Phosphoprotein 8 (MPP8) is highly expressed in ESCs and germline cells. Although evidence of a cooperation between H3K9 KMTs and MPP8 in committed cells has emerged, the interplay between H3K9 methylation writers and MPP8 in ESCs remains elusive. -

Deep Multiomics Profiling of Brain Tumors Identifies Signaling Networks
ARTICLE https://doi.org/10.1038/s41467-019-11661-4 OPEN Deep multiomics profiling of brain tumors identifies signaling networks downstream of cancer driver genes Hong Wang 1,2,3, Alexander K. Diaz3,4, Timothy I. Shaw2,5, Yuxin Li1,2,4, Mingming Niu1,4, Ji-Hoon Cho2, Barbara S. Paugh4, Yang Zhang6, Jeffrey Sifford1,4, Bing Bai1,4,10, Zhiping Wu1,4, Haiyan Tan2, Suiping Zhou2, Laura D. Hover4, Heather S. Tillman 7, Abbas Shirinifard8, Suresh Thiagarajan9, Andras Sablauer 8, Vishwajeeth Pagala2, Anthony A. High2, Xusheng Wang 2, Chunliang Li 6, Suzanne J. Baker4 & Junmin Peng 1,2,4 1234567890():,; High throughput omics approaches provide an unprecedented opportunity for dissecting molecular mechanisms in cancer biology. Here we present deep profiling of whole proteome, phosphoproteome and transcriptome in two high-grade glioma (HGG) mouse models driven by mutated RTK oncogenes, PDGFRA and NTRK1, analyzing 13,860 proteins and 30,431 phosphosites by mass spectrometry. Systems biology approaches identify numerous master regulators, including 41 kinases and 23 transcription factors. Pathway activity computation and mouse survival indicate the NTRK1 mutation induces a higher activation of AKT down- stream targets including MYC and JUN, drives a positive feedback loop to up-regulate multiple other RTKs, and confers higher oncogenic potency than the PDGFRA mutation. A mini-gRNA library CRISPR-Cas9 validation screening shows 56% of tested master regulators are important for the viability of NTRK-driven HGG cells, including TFs (Myc and Jun) and metabolic kinases (AMPKa1 and AMPKa2), confirming the validity of the multiomics inte- grative approaches, and providing novel tumor vulnerabilities.