Deoxyhypusine Synthase As an Antimalarial Target
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Yeast Genome Gazetteer P35-65
gazetteer Metabolism 35 tRNA modification mitochondrial transport amino-acid metabolism other tRNA-transcription activities vesicular transport (Golgi network, etc.) nitrogen and sulphur metabolism mRNA synthesis peroxisomal transport nucleotide metabolism mRNA processing (splicing) vacuolar transport phosphate metabolism mRNA processing (5’-end, 3’-end processing extracellular transport carbohydrate metabolism and mRNA degradation) cellular import lipid, fatty-acid and sterol metabolism other mRNA-transcription activities other intracellular-transport activities biosynthesis of vitamins, cofactors and RNA transport prosthetic groups other transcription activities Cellular organization and biogenesis 54 ionic homeostasis organization and biogenesis of cell wall and Protein synthesis 48 plasma membrane Energy 40 ribosomal proteins organization and biogenesis of glycolysis translation (initiation,elongation and cytoskeleton gluconeogenesis termination) organization and biogenesis of endoplasmic pentose-phosphate pathway translational control reticulum and Golgi tricarboxylic-acid pathway tRNA synthetases organization and biogenesis of chromosome respiration other protein-synthesis activities structure fermentation mitochondrial organization and biogenesis metabolism of energy reserves (glycogen Protein destination 49 peroxisomal organization and biogenesis and trehalose) protein folding and stabilization endosomal organization and biogenesis other energy-generation activities protein targeting, sorting and translocation vacuolar and lysosomal -

Anti-Inflammatory Role of Curcumin in LPS Treated A549 Cells at Global Proteome Level and on Mycobacterial Infection
Anti-inflammatory Role of Curcumin in LPS Treated A549 cells at Global Proteome level and on Mycobacterial infection. Suchita Singh1,+, Rakesh Arya2,3,+, Rhishikesh R Bargaje1, Mrinal Kumar Das2,4, Subia Akram2, Hossain Md. Faruquee2,5, Rajendra Kumar Behera3, Ranjan Kumar Nanda2,*, Anurag Agrawal1 1Center of Excellence for Translational Research in Asthma and Lung Disease, CSIR- Institute of Genomics and Integrative Biology, New Delhi, 110025, India. 2Translational Health Group, International Centre for Genetic Engineering and Biotechnology, New Delhi, 110067, India. 3School of Life Sciences, Sambalpur University, Jyoti Vihar, Sambalpur, Orissa, 768019, India. 4Department of Respiratory Sciences, #211, Maurice Shock Building, University of Leicester, LE1 9HN 5Department of Biotechnology and Genetic Engineering, Islamic University, Kushtia- 7003, Bangladesh. +Contributed equally for this work. S-1 70 G1 S 60 G2/M 50 40 30 % of cells 20 10 0 CURI LPSI LPSCUR Figure S1: Effect of curcumin and/or LPS treatment on A549 cell viability A549 cells were treated with curcumin (10 µM) and/or LPS or 1 µg/ml for the indicated times and after fixation were stained with propidium iodide and Annexin V-FITC. The DNA contents were determined by flow cytometry to calculate percentage of cells present in each phase of the cell cycle (G1, S and G2/M) using Flowing analysis software. S-2 Figure S2: Total proteins identified in all the three experiments and their distribution betwee curcumin and/or LPS treated conditions. The proteins showing differential expressions (log2 fold change≥2) in these experiments were presented in the venn diagram and certain number of proteins are common in all three experiments. -

Supplementary Table S4. FGA Co-Expressed Gene List in LUAD
Supplementary Table S4. FGA co-expressed gene list in LUAD tumors Symbol R Locus Description FGG 0.919 4q28 fibrinogen gamma chain FGL1 0.635 8p22 fibrinogen-like 1 SLC7A2 0.536 8p22 solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 DUSP4 0.521 8p12-p11 dual specificity phosphatase 4 HAL 0.51 12q22-q24.1histidine ammonia-lyase PDE4D 0.499 5q12 phosphodiesterase 4D, cAMP-specific FURIN 0.497 15q26.1 furin (paired basic amino acid cleaving enzyme) CPS1 0.49 2q35 carbamoyl-phosphate synthase 1, mitochondrial TESC 0.478 12q24.22 tescalcin INHA 0.465 2q35 inhibin, alpha S100P 0.461 4p16 S100 calcium binding protein P VPS37A 0.447 8p22 vacuolar protein sorting 37 homolog A (S. cerevisiae) SLC16A14 0.447 2q36.3 solute carrier family 16, member 14 PPARGC1A 0.443 4p15.1 peroxisome proliferator-activated receptor gamma, coactivator 1 alpha SIK1 0.435 21q22.3 salt-inducible kinase 1 IRS2 0.434 13q34 insulin receptor substrate 2 RND1 0.433 12q12 Rho family GTPase 1 HGD 0.433 3q13.33 homogentisate 1,2-dioxygenase PTP4A1 0.432 6q12 protein tyrosine phosphatase type IVA, member 1 C8orf4 0.428 8p11.2 chromosome 8 open reading frame 4 DDC 0.427 7p12.2 dopa decarboxylase (aromatic L-amino acid decarboxylase) TACC2 0.427 10q26 transforming, acidic coiled-coil containing protein 2 MUC13 0.422 3q21.2 mucin 13, cell surface associated C5 0.412 9q33-q34 complement component 5 NR4A2 0.412 2q22-q23 nuclear receptor subfamily 4, group A, member 2 EYS 0.411 6q12 eyes shut homolog (Drosophila) GPX2 0.406 14q24.1 glutathione peroxidase -

A Novel Mouse Model for Inhibition of DOHH-Mediated Hypusine Modification Reveals a Crucial Function in Embryonic Development, P
© 2014. Published by The Company of Biologists Ltd | Disease Models & Mechanisms (2014) 7, 963-976 doi:10.1242/dmm.014449 RESEARCH ARTICLE A novel mouse model for inhibition of DOHH-mediated hypusine modification reveals a crucial function in embryonic development, proliferation and oncogenic transformation Henning Sievert1, Nora Pällmann1,2,*, Katharine K. Miller3,*, Irm Hermans-Borgmeyer3, Simone Venz4, Ataman Sendoel5,6, Michael Preukschas1, Michaela Schweizer3, Steffen Boettcher6, P. Christoph Janiesch3, Thomas Streichert7, Reinhard Walther4, Michael O. Hengartner5, Markus G. Manz6, Tim H. Brümmendorf8, Carsten Bokemeyer1, Melanie Braig1, Joachim Hauber2, Kent E. Duncan3 and Stefan Balabanov1,6,‡ ABSTRACT factor 5A (eIF5A), represents an essential mechanism in the control The central importance of translational control by post-translational of proliferation of eukaryotic cells (Cooper et al., 1982). This modification has spurred major interest in regulatory pathways that modification leads to the activation of eIF5A and is mediated by control translation. One such pathway uniquely adds hypusine to deoxyhypusine synthase (DHS), which catalyses the transfer of a 4- eukaryotic initiation factor 5A (eIF5A), and thereby affects protein aminobutyl moiety of spermidine to the ε-amino group of Lys50 to synthesis and, subsequently, cellular proliferation through an form an intermediate residue, deoxyhypusine (Dhp50) (Park et al., unknown mechanism. Using a novel conditional knockout mouse 1981). Subsequently, deoxyhypusine hydroxylase (DOHH) -

12) United States Patent (10
US007635572B2 (12) UnitedO States Patent (10) Patent No.: US 7,635,572 B2 Zhou et al. (45) Date of Patent: Dec. 22, 2009 (54) METHODS FOR CONDUCTING ASSAYS FOR 5,506,121 A 4/1996 Skerra et al. ENZYME ACTIVITY ON PROTEIN 5,510,270 A 4/1996 Fodor et al. MICROARRAYS 5,512,492 A 4/1996 Herron et al. 5,516,635 A 5/1996 Ekins et al. (75) Inventors: Fang X. Zhou, New Haven, CT (US); 5,532,128 A 7/1996 Eggers Barry Schweitzer, Cheshire, CT (US) 5,538,897 A 7/1996 Yates, III et al. s s 5,541,070 A 7/1996 Kauvar (73) Assignee: Life Technologies Corporation, .. S.E. al Carlsbad, CA (US) 5,585,069 A 12/1996 Zanzucchi et al. 5,585,639 A 12/1996 Dorsel et al. (*) Notice: Subject to any disclaimer, the term of this 5,593,838 A 1/1997 Zanzucchi et al. patent is extended or adjusted under 35 5,605,662 A 2f1997 Heller et al. U.S.C. 154(b) by 0 days. 5,620,850 A 4/1997 Bamdad et al. 5,624,711 A 4/1997 Sundberg et al. (21) Appl. No.: 10/865,431 5,627,369 A 5/1997 Vestal et al. 5,629,213 A 5/1997 Kornguth et al. (22) Filed: Jun. 9, 2004 (Continued) (65) Prior Publication Data FOREIGN PATENT DOCUMENTS US 2005/O118665 A1 Jun. 2, 2005 EP 596421 10, 1993 EP 0619321 12/1994 (51) Int. Cl. EP O664452 7, 1995 CI2O 1/50 (2006.01) EP O818467 1, 1998 (52) U.S. -

Supplementary Table S3. Core Genome Chromosomal Positioning
Supplementary Table S3. Core genome chromosomal positioning % Mean distance % distance standard Pyrococcus abyssi P. abyssi ortholog definition P. abyssi P. -
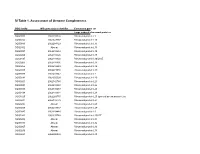
SI Table 1. Assessment of Genome Completeness
SI Table 1. Assessment of Genome Completeness COG family IMG gene object identifier Conserved gene set Large subunit ribosomal proteins COG0081 2062288324 Ribosomal protein L1 COG0244 2062347387 Ribosomal protein L10 COG0080 2062288323 Ribosomal protein L11 COG0102 Absent Ribosomal protein L13 COG0093 2062418832 Ribosomal protein L14 COG0200 2062418826 Ribosomal protein L15 COG0197 2062418838 Ribosomal protein L16/L10E COG0203 2062418836 Ribosomal protein L17 COG0256 2062418829 Ribosomal protein L18 COG0335 2062273558 Ribosomal protein L19 COG0090 2062418842 Ribosomal protein L2 COG0292 2062350539 Ribosomal protein L20 COG0261 2062142780 Ribosomal protein L21 COG0091 2062418840 Ribosomal protein L22 COG0089 2062138283 Ribosomal protein L23 COG0198 2062418834 Ribosomal protein L24 COG1825 2062269715 Ribosomal protein L25 (general stress protein Ctc) COG0211 2062142779 Ribosomal protein L27 COG0227 Absent Ribosomal protein L28 COG0255 2062418837 Ribosomal protein L29 COG0087 2062154483 Ribosomal protein L3 COG1841 2062335748 Ribosomal protein L30/L7E COG0254 Absent Ribosomal protein L31 COG0333 Absent Ribosomal protein L32 COG0267 Absent Ribosomal protein L33 COG0230 Absent Ribosomal protein L34 COG0291 2062350538 Ribosomal protein L35 COG0257 Absent Ribosomal protein L36 COG0088 2062138282 Ribosomal protein L4 COG0094 2062418833 Ribosomal protein L5 COG0097 2062418830 Ribosomal protein L6P/L9E COG0222 2062288326 Ribosomal protein L7/L12 COG0359 2062209880 Ribosomal protein L9 Small subunit ribosomal proteins COG0539 Absent Ribosomal protein -

Cloning and Expression of Deoxyhypusine Synthase from Plasmodium Vivax and Theileria Parva As an Approach for Target Evaluation in Anti-Parasitic Chemotherapy
Cloning and expression of deoxyhypusine synthase from Plasmodium vivax and Theileria parva as an approach for target evaluation in anti-parasitic chemotherapy Dissertation zur Erlangung des Doktorgrades (Dr. rer. nat.) der Mathematisch-Naturwissenschaftlichen Fakultät der Rheinischen Friedrich-Wilhelms-Universität Bonn vorgelegt von James Thuo Njuguna aus Ol Kalou,Kenya Bonn (2009) I Angefertigt mit Genehmigung der Mathematisch-Naturwissenschaftlichen Fakultät der Rheinischen Friedrich-Wilhelms-Universität Bonn 1. Gutachter: Prof. Dr. Annette.E. Kaiser 2. Gutachter: Prof. Dr. Christa E. Müller Tag der Promotion: (31-09-2009) II Acknowledgments This thesis has come to realization because of the support and mentoring of my supervisors Prof. Dr. Annette Kaiser and Prof. Dr Christa Mueller. Prof. Dr Kaiser led me to this area of study and has kept me focused on our scientific goals even when faced with apparently insurmountable challenges. My co-supervisor Prof. Dr. Prof. Christa Mueller has been of great help and support during the last phase of the study. To both I express my deepest gratitude. This Thesis was only possible because of the logistical support by Peter Croll and his institute the Bonn International Center for Conversion (BICC); I will always be indebted to him for this support and for his ideas on positive human development. I am very thankful to Prof. Dr. Achim Hörauf for hosting my study at the Institute for Medical Microbiology, Immunology and Parasitology when I first got started and for his continued interest in our progress. I would like to also thank Prof. Dr. U. Holzgrabe for availing to me the piperidones evaluated in this Thesis, Dr. -

Autocrine IFN Signaling Inducing Profibrotic Fibroblast Responses By
Downloaded from http://www.jimmunol.org/ by guest on September 23, 2021 Inducing is online at: average * The Journal of Immunology , 11 of which you can access for free at: 2013; 191:2956-2966; Prepublished online 16 from submission to initial decision 4 weeks from acceptance to publication August 2013; doi: 10.4049/jimmunol.1300376 http://www.jimmunol.org/content/191/6/2956 A Synthetic TLR3 Ligand Mitigates Profibrotic Fibroblast Responses by Autocrine IFN Signaling Feng Fang, Kohtaro Ooka, Xiaoyong Sun, Ruchi Shah, Swati Bhattacharyya, Jun Wei and John Varga J Immunol cites 49 articles Submit online. Every submission reviewed by practicing scientists ? is published twice each month by Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts http://jimmunol.org/subscription Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html http://www.jimmunol.org/content/suppl/2013/08/20/jimmunol.130037 6.DC1 This article http://www.jimmunol.org/content/191/6/2956.full#ref-list-1 Information about subscribing to The JI No Triage! Fast Publication! Rapid Reviews! 30 days* Why • • • Material References Permissions Email Alerts Subscription Supplementary The Journal of Immunology The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2013 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. This information is current as of September 23, 2021. The Journal of Immunology A Synthetic TLR3 Ligand Mitigates Profibrotic Fibroblast Responses by Inducing Autocrine IFN Signaling Feng Fang,* Kohtaro Ooka,* Xiaoyong Sun,† Ruchi Shah,* Swati Bhattacharyya,* Jun Wei,* and John Varga* Activation of TLR3 by exogenous microbial ligands or endogenous injury-associated ligands leads to production of type I IFN. -

Discovery of High Affinity Receptors for Dityrosine Through Inverse Virtual Screening and Docking and Molecular Dynamics
Article Discovery of High Affinity Receptors for Dityrosine through Inverse Virtual Screening and Docking and Molecular Dynamics Fangfang Wang 1,*,†, Wei Yang 2,3,† and Xiaojun Hu 1,* 1 School of Life Science, Linyi University, Linyi 276000, China; [email protected] 2 Department of Microbiology, Biomedicine Discovery Institute, Monash University, Clayton, VIC 3800, Australia, [email protected] 3 Arieh Warshel Institute of Computational Biology, the Chinese University of Hong Kong, 2001 Longxiang Road, Longgang District, Shenzhen 518000, China * Corresponding author: [email protected] † These authors contributed equally to this work. Received: 09 December 2018; Accepted: 23 December 2018; Published: date Table S1. Docking affinity scores for cis-dityrosine binding to binding proteins. Target name PDB/UniProtKB Type Affinity (kcal/mol) Galectin-1 1A78/P56217 Lectin -6.2±0.0 Annexin III 1AXN/P12429 Calcium/phospholipid Binding Protein -7.5±0.0 Calmodulin 1CTR/P62158 Calcium Binding Protein -5.8±0.0 Seminal Plasma Protein Pdc-109 1H8P/P02784 Phosphorylcholine Binding Protein -6.6±0.0 Annexin V 1HAK/P08758 Calcium/phospholipid Binding -7.4±0.0 Alpha 1 antitrypsin 1HP7/P01009 Protein Binding -7.6±0.0 Histidine-Binding Protein 1HSL/P0AEU0 Binding Protein -6.3±0.0 Intestinal Fatty Acid Binding Protein 1ICN/P02693 Binding Protein(fatty Acid) -9.1±0.0* Migration Inhibitory Factor-Related Protein 14 1IRJ/P06702 Metal Binding Protein -7.0±0.0 Lysine-, Arginine-, Ornithine-Binding Protein 1LST/P02911 Amino Acid Binding Protein -6.5±0.0 -

Halfway to Hypusine—Structural Basis for Substrate Recognition by Human
biomolecules Article Half Way to Hypusine—Structural Basis for Substrate Recognition by Human Deoxyhypusine Synthase El˙zbietaW ˛ator , Piotr Wilk and Przemysław Grudnik * Malopolska Centre of Biotechnology, Jagiellonian University, ul. Gronostajowa 7a, 30-387 Krakow, Poland; [email protected] (E.W.); [email protected] (P.W.) * Correspondence: [email protected] Received: 30 January 2020; Accepted: 26 March 2020; Published: 30 March 2020 Abstract: Deoxyhypusine synthase (DHS) is a transferase enabling the formation of deoxyhypusine, which is the first, rate-limiting step of a unique post-translational modification: hypusination. DHS catalyses the transfer of a 4-aminobutyl moiety of polyamine spermidine to a specific lysine of eukaryotic translation factor 5A (eIF5A) precursor in a nicotinamide adenine dinucleotide (NAD)-dependent manner. This modification occurs exclusively on one protein, eIF5A, and it is essential for cell proliferation. Malfunctions of the hypusination pathway, including those caused by mutations within the DHS encoding gene, are associated with conditions such as cancer or neurodegeneration. Here, we present a series of high-resolution crystal structures of human DHS. Structures were determined as the apoprotein, as well as ligand-bound states at high-resolutions ranging from 1.41 to 1.69 Å. By solving DHS in complex with its natural substrate spermidine (SPD), we identified the mode of substrate recognition. We also observed that other polyamines, namely spermine (SPM) and putrescine, bind DHS in a similar manner as SPD. Moreover, we performed activity assays showing that SPM could to some extent serve as an alternative DHS substrate. In contrast to previous studies, we demonstrate that no conformational changes occur in the DHS structure upon spermidine-binding. -
PDF) Tributions Package from CPAN Was Used for Obtaining P-Values
A Measure of the Promiscuity of Proteins and Characteristics of Residues in the Vicinity of the Catalytic Site That Regulate Promiscuity Sandeep Chakraborty, Basuthkar J. Rao* Department of Biological Sciences, Tata Institute of Fundamental Research, Mumbai, India, Abstract Promiscuity, the basis for the evolution of new functions through ‘tinkering’ of residues in the vicinity of the catalytic site, is yet to be quantitatively defined. We present a computational method Promiscuity IndicesEstimator (PROMISE) - based on signatures derived from the spatial and electrostatic properties of the catalytic residues, to estimate the promiscuity (PromIndex) of proteins with known active site residues and 3D structure. PromIndex reflects the number of different active site signatures that have congruent matches in close proximity of its native catalytic site, the quality of the matches and difference in the enzymatic activity. Promiscuity in proteins is observed to follow a lognormal distribution (m = 0.28, s = 1.1 reduced chi-square = 3.0E-5). The PROMISE predicted promiscuous functions in any protein can serve as the starting point for directed evolution experiments. PROMISE ranks carboxypeptidase A and ribonuclease A amongst the more promiscuous proteins. We have also investigated the properties of the residues in the vicinity of the catalytic site that regulates its promiscuity. Linear regression establishes a weak correlation (R2,0.1) between certain properties of the residues (charge, polar, etc) in the neighborhood of the catalytic residues and PromIndex. A stronger relationship states that most proteins with high promiscuity have high percentages of charged and polar residues within a radius of 3 A˚ of the catalytic site, which is validated using one-tailed hypothesis tests (P-values,0.05).