Sequence-Engineered Mrna Without Chemical Nucleoside Modifications Enables an Effective Protein Therapy in Large Animals
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Split Deoxyribozyme Probe for Efficient Detection of Highly Structured RNA Targets
University of Central Florida STARS Honors Undergraduate Theses UCF Theses and Dissertations 2018 Split Deoxyribozyme Probe For Efficient Detection of Highly Structured RNA Targets Sheila Raquel Solarez University of Central Florida Part of the Biochemistry Commons, and the Biology Commons Find similar works at: https://stars.library.ucf.edu/honorstheses University of Central Florida Libraries http://library.ucf.edu This Open Access is brought to you for free and open access by the UCF Theses and Dissertations at STARS. It has been accepted for inclusion in Honors Undergraduate Theses by an authorized administrator of STARS. For more information, please contact [email protected]. Recommended Citation Solarez, Sheila Raquel, "Split Deoxyribozyme Probe For Efficient Detection of Highly Structured RNA Targets" (2018). Honors Undergraduate Theses. 311. https://stars.library.ucf.edu/honorstheses/311 SPLIT DEOXYRIBOZYME PROBE FOR EFFICIENT DETECTION OF HIGHLY STRUCTURED RNA TARGETS By SHEILA SOLAREZ A thesis submitted in partial fulfillment of the requirements for the Honors in the Major Program in Biological Sciences in the College of Sciences and the Burnett Honors College at the University of Central Florida Orlando, Florida Spring Term, 2018 Thesis Chair: Yulia Gerasimova, PhD ABSTRACT Transfer RNAs (tRNAs) are known for their role as adaptors during translation of the genetic information and as regulators for gene expression; uncharged tRNAs regulate global gene expression in response to changes in amino acid pools in the cell. Aminoacylated tRNAs play a role in non-ribosomal peptide bond formation, post-translational protein labeling, modification of phospholipids in the cell membrane, and antibiotic biosynthesis. [1] tRNAs have a highly stable structure that can present a challenge for their detection using conventional techniques. -

Mrna Vaccine Era—Mechanisms, Drug Platform and Clinical Prospection
International Journal of Molecular Sciences Review mRNA Vaccine Era—Mechanisms, Drug Platform and Clinical Prospection 1, 1, 2 1,3, Shuqin Xu y, Kunpeng Yang y, Rose Li and Lu Zhang * 1 State Key Laboratory of Genetic Engineering, Institute of Genetics, School of Life Science, Fudan University, Shanghai 200438, China; [email protected] (S.X.); [email protected] (K.Y.) 2 M.B.B.S., School of Basic Medical Sciences, Peking University Health Science Center, Beijing 100191, China; [email protected] 3 Shanghai Engineering Research Center of Industrial Microorganisms, Shanghai 200438, China * Correspondence: [email protected]; Tel.: +86-13524278762 These authors contributed equally to this work. y Received: 30 July 2020; Accepted: 30 August 2020; Published: 9 September 2020 Abstract: Messenger ribonucleic acid (mRNA)-based drugs, notably mRNA vaccines, have been widely proven as a promising treatment strategy in immune therapeutics. The extraordinary advantages associated with mRNA vaccines, including their high efficacy, a relatively low severity of side effects, and low attainment costs, have enabled them to become prevalent in pre-clinical and clinical trials against various infectious diseases and cancers. Recent technological advancements have alleviated some issues that hinder mRNA vaccine development, such as low efficiency that exist in both gene translation and in vivo deliveries. mRNA immunogenicity can also be greatly adjusted as a result of upgraded technologies. In this review, we have summarized details regarding the optimization of mRNA vaccines, and the underlying biological mechanisms of this form of vaccines. Applications of mRNA vaccines in some infectious diseases and cancers are introduced. It also includes our prospections for mRNA vaccine applications in diseases caused by bacterial pathogens, such as tuberculosis. -

Expanding the Genetic Code Lei Wang and Peter G
Reviews P. G. Schultz and L. Wang Protein Science Expanding the Genetic Code Lei Wang and Peter G. Schultz* Keywords: amino acids · genetic code · protein chemistry Angewandte Chemie 34 2005 Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim DOI: 10.1002/anie.200460627 Angew. Chem. Int. Ed. 2005, 44,34–66 Angewandte Protein Science Chemie Although chemists can synthesize virtually any small organic molecule, our From the Contents ability to rationally manipulate the structures of proteins is quite limited, despite their involvement in virtually every life process. For most proteins, 1. Introduction 35 modifications are largely restricted to substitutions among the common 20 2. Chemical Approaches 35 amino acids. Herein we describe recent advances that make it possible to add new building blocks to the genetic codes of both prokaryotic and 3. In Vitro Biosynthetic eukaryotic organisms. Over 30 novel amino acids have been genetically Approaches to Protein encoded in response to unique triplet and quadruplet codons including Mutagenesis 39 fluorescent, photoreactive, and redox-active amino acids, glycosylated 4. In Vivo Protein amino acids, and amino acids with keto, azido, acetylenic, and heavy-atom- Mutagenesis 43 containing side chains. By removing the limitations imposed by the existing 20 amino acid code, it should be possible to generate proteins and perhaps 5. An Expanded Code 46 entire organisms with new or enhanced properties. 6. Outlook 61 1. Introduction The genetic codes of all known organisms specify the same functional roles to amino acid residues in proteins. Selectivity 20 amino acid building blocks. These building blocks contain a depends on the number and reactivity (dependent on both limited number of functional groups including carboxylic steric and electronic factors) of a particular amino acid side acids and amides, a thiol and thiol ether, alcohols, basic chain. -
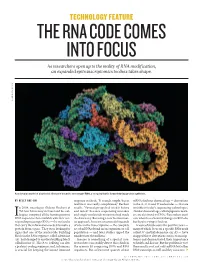
THE RNA CODE COMES INTO FOCUS As Researchers Open up to the Reality of RNA Modification, an Expanded Epitranscriptomics Toolbox Takes Shape
TECHNOLOGY FEATURE THE RNA CODE COMES INTO FOCUS As researchers open up to the reality of RNA modification, an expanded epitranscriptomics toolbox takes shape. LAGUNA DESIGN/SPL LAGUNA A molecular model of a bacterial ribosome bound to messenger RNA, a complex that is formed during protein synthesis. BY KELLY RAE CHI response in check. “It sounds simple, but in mRNAs harbour chemical tags — decorations real life it was really complicated,” Rechavi to the A, C, G and U nucleotides — that are n 2004, oncologist Gideon Rechavi at recalls. “Several groups had tried it before invisible to today’s sequencing technologies. Tel Aviv University in Israel and his col- and failed” because sequencing mistakes (Similar chemical tags, called epigenetic mark- leagues compared all the human genomic and single-nucleotide mutations had made ers, are also found on DNA.) Researchers aren’t IDNA sequences then available with their cor- the data noisy. But using a new bioinformat- sure what these chemical changes in RNA do, responding messenger RNAs — the molecules ics approach, his team uncovered thousands but they’re trying to find out. that carry the information needed to make a of sites in the transcriptome — the complete A wave of studies over the past five years — protein from a gene. They were looking for set of mRNAs found in an organism or cell many of which focus on a specific RNA mark signs that one of the nucleotide building population — and later studies upped the called N6-methyladenosine (m6A) — have blocks in the RNA sequence, called adenosine number into the millions1. -

Mrna, Rrna and Trna Types of RNA: Mrna, Rrna and Trna
Types of RNA: mRNA, rRNA and tRNA Types of RNA: mRNA, rRNA and tRNA By Susha Cheriyedath, M.Sc. Reviewed by Michael Greenwood, M.Sc. RNA or ribonucleic acid is a polymer of nucleotides that is made up of a ribose sugar, a phosphate, and bases such as adenine, guanine, cytosine, and uracil. It plays a crucial role in gene expression by acting as the intermediate between the genetic information encoded by DNA and proteins. Designua | Shutterstock RNA has a structure very similar to that of DNA. The key difference in RNA structure is that the ribose sugar in RNA possesses a hydroxyl (OH) group that is absent in DNA. Types of RNA In both prokaryotes and eukaryotes, there are three main types of RNA – messenger RNA (mRNA), ribosomal RNA (rRNA), and transfer RNA (tRNA). These 3 types of RNA are discussed below. P Saved from URL: https://www.news-medical.net/life-sciences/-Types-of-RNA-mRNA-rRNA-and-tRNA.aspx 1/5 Types of RNA: mRNA, rRNA and tRNA Messenger RNA (mRNA) mRNA accounts for just 5% of the total RNA in the cell. mRNA is the most heterogeneous of the 3 types of RNA in terms of both base sequence and size. It carries complimentary genetic code copied, from DNA during transcription, in the form of triplets of nucleotides called codons. Each codon specifies a particular amino acid, though one amino acid may be coded for by many different codons. Although there are 64 possible codons or triplet bases in the genetic code, only 20 of them represent amino acids. -

Transcription Study Guide This Study Guide Is a Written Version of the Material You Have Seen Presented in the Transcription Unit
Transcription Study Guide This study guide is a written version of the material you have seen presented in the transcription unit. The cell’s DNA contains the instructions for carrying out the work of the cell. These instructions are used by the cell’s protein-making machinery to create proteins. If the cell’s DNA were directly read by the protein-making machinery, however, it could be damaged and the process would be slow and cumbersome. The cell avoids this problem by copying genetic information from its DNA into an intermediate called messenger RNA (mRNA). It is this mRNA that is read by the cell’s protein-making machinery. This process is called transcription. Components In this section you will be introduced to the components involved in the process of RNA synthesis, called transcription. This process requires an enzyme that uses many nucleotide bases to copy the instructions present in DNA into an intermediate messenger RNA molecule. RNA What is RNA? · Like DNA, RNA is a polymer made up of nucleotides. · Unlike DNA, which is composed of two strands of nucleotides twisted together, RNA is single-stranded. It can also sometimes fold into complex three-dimensional structures. · RNA contains the same nucleotides as DNA, with the substitution of uraciluridine (U) for thymidine (T). · RNA is chemically different from DNA so that the cell can easily tell the two apart. · In this animation, you will see one type of RNA, messenger RNA, being put together. · There are three types of RNA: mRNA, which you will read more about; tRNA, which is used in the translation process, and rRNA, which acts as a structural element in the ribosome (a translation component). -

Isolation of RNA with Properties of Messenger RNA from Cerebral Polyribosomes* Claire E
Proceedings of the National Academy of Sciences Vol. 67, No. 2, pp. 644-651, October 1970 Isolation of RNA with Properties of Messenger RNA from Cerebral Polyribosomes* Claire E. Zomzelyt, Sidney Roberts, and Susan Peache DEPARTMENT OF BIOLOGICAL CHEMISTRY, SCHOOL OF MEDICINE, AND THE BRAIN RESEARCH INSTI- TUTE, UNIVERSITY OF CALIFORNIA CENTER FOR THE HEALTH SCIENCES, LOS ANGELES, CALIFORNIA 90024 Communicated by H. W. Magoun, July 24, 1970 Abstract. RNA which dissociated from purified cerebral polyribosomes of adult rats in the presence of EDTA was isolated by fractionation in a dis- continuous sucrose gradient. The yield was 2% of the total polyribosomal RNA. The base composition resembled the complementary values for rat DNA and was very different from base compositions of ribosomal RNA and transfer RNA. This RNA fraction contained a large proportion of molecules which were rapidly labeled in vivo and hybridized to homologous DNA. The polyribosomal RNA preparation also exhibited high template activity in a cerebral cell-free system which had previously been stripped of the capacity to incorporate amino acids in the absence of added messenger RNA (mRNA). Sedimentation analysis revealed only two peaks, with coefficients of approximately 8 S and 16 S. The data indicate that RNA with the properties of mRNA can be selectively isolated from cerebral polyribosomes under mild conditions which avoid degradation. Numerous studies support the concept that alterations in RNA and protein synthesis are associated with information transfer and storage in the central nervous system. Activation of the synthesis of specific proteins is suggested by the reports that new RNA with altered base composition, presumably mRNA, is formed during the acquisition phase of learning. -

Why We Should Take a Second Look at RNA Technology
RNA THERAPEUTICS RNA Modulation: Gabriele Campi Why we should take a second look at RNA technology As a scientist first, but also as an investor, I have always hereditary ATTR amyloidosis with polyneuropathy, an believed in the potential of RNA molecules as therapeutics. autosomal dominant neurodegenerative disease. In 2018 While most current drugs aim to stop disease by modulating patisiran (Onpattro) was approved by the FDA. It is existing proteins, RNA molecules operate one step earlier. delivered to cells by lipid nanoparticles rather than GalNAc In the case of RNA interference (RNAi), they modulate the (N-Acetylgalactoseamine-siRNA conjugates). genes involved in different pathological processes. The RNA Since the end of 2018, investors have made a cautious molecules, small interfering RNA and microRNA, both return to the RNA therapy space with a targeted funding degrade messenger RNA (mRNA) and prevent it from being of existing RNA companies. In parallel, new companies are translated into proteins. entering the arena. An unexpected boost to the industry has I am the co-founder of the investment company AurorA come from the development and approval of the first mRNA Science, which recently led a Series B financing round for the vaccines to treat Covid-19 from Pfizer/BioNTech and Moderna. Dutch company InteRNA Technologies, which is developing I have always believed in the potency of RNA interference RNA therapeutics for the treatment of advanced solid and modulation techniques that allow a researcher to target tumours. This is an ambitious project, but with a potential specific messenger RNAs through the use of siRNAs, or for success. -

Pfizer-Biontech COVID-19 Vaccine)
Nucleoside-modified messenger RNA (modmRNA) of SARS-CoV-2 Suspension for injection (Pfizer-BioNTech COVID-19 Vaccine) U.S. Trade Names Pfizer-BioNTech COVID-19 The list of names may not include all products that are available on the market. What is this medicine? SARS COVID-19 VACCINE (SARZ koh-vid 19 vak SEEN) is a vaccine used to reduce the risk of getting COVID-19. This vaccine does not treat COVID-19. There is no FDA-approved vaccine to prevent COVID-19. The FDA has authorized the emergency use of this vaccine during the COVID-19 pandemic. This medicine may be used for other purposes; ask your health care provider or pharmacist if you have questions. What should I tell my health care provider before I take this medicine? They need to know if you have any of these conditions: any allergies bleeding disorder fever or infection immune system problems recent or upcoming vaccine including previous COVID-19 vaccine an unusual or allergic reaction to COVID-19 vaccine, other drugs, foods, dyes, or preservatives pregnant or trying to get pregnant breast-feeding How should I use this medicine? This vaccine is injected into a muscle. It is given by a health care professional. A copy of the Fact Sheet for Recipients and Caregivers will be given before each vaccination. Be sure to read this information carefully each time. This sheet may change frequently. Talk to your health care provider about the use of this drug in children. Special care may be needed. Overdosage: If you think you have taken too much of this medicine contact a poison control center or emergency room at once. -

Ribonucleic Acid (RNA)
AccessScience from McGraw-Hill Education Page 1 of 11 www.accessscience.com Ribonucleic acid (RNA) Contributed by: Michael W. Gray, Ann L. Beyer Publication year: 2014 One of the two major classes of nucleic acid, mainly involved in translating the genetic information carried in deoxyribonucleic acid (DNA) into proteins. Various types of ribonucleic acids (RNAs) [see table] function in protein synthesis: transfer RNAs (tRNAs) and ribosomal RNAs (rRNAs) function in the synthesis of all proteins, whereas messenger RNAs (mRNAs) are a diverse set, with each member specifically directing the synthesis of one protein. Messenger RNA is the intermediate in the usual biological pathway of DNA → RNA → protein. However, RNA is a very versatile molecule. Other types of RNA serve other important functions for cells and viruses, such as the involvement of small nuclear RNAs (snRNAs) in mRNA splicing. In some cases, RNA performs functions typically considered DNA-like, such as serving as the genetic material for certain viruses, or roles typically carried out by proteins, such as RNA enzymes or ribozymes. See also: DEOXYRIBONUCLEIC ACID (DNA); NUCLEIC ACID. Structure and synthesis RNA is a linear polymer of four different nucleotides (Fig. 1). Each nucleotide is composed of three parts: a five-carbon sugar known as ribose; a phosphate group; and one of four bases attached to each ribose, that is, adenine (A), cytosine (C), guanine (G), or uracil (U). The combination of base and sugar constitutes a nucleoside. The structure of RNA is basically a repeating chain of ribose and phosphate moieties, with one of the four bases attached to each ribose. -

RNA and ITS STRUCTURE, FUNCTION and TYPES with The
RNA AND ITS STRUCTURE, FUNCTION AND TYPES With the discovery of the molecular structure of the DNA double helix in 1953, researchers turned to the structure of ribonucleic acid (RNA) as the next critical puzzle to be solved on the road to understanding the molecular basis of life. Ribonucleic acid (RNA) is a type of molecule that consists of a long chain of nucleotide units. Each nucleotide consists of a nitrogenous base, a ribose sugar, and a phosphate. RNA is very similar to DNA, but differs in a few important structural details: in the cell, RNA is usually single-stranded, while DNA is usually double-stranded; RNA nucleotides contain ribose while DNA contains deoxyribose (a type of ribose that lacks one oxygen atom); and RNA has the base uracil rather than thymine that is present in DNA. RNA is transcribed from DNA by enzymes called RNA polymerases and is generally further processed by other enzymes. RNA is central to the synthesis of proteins. Here, a type of RNA called messenger RNA carries information from DNA to structures called ribosomes. These ribosomes are made from proteins and ribosomal RNAs, which come together to form a molecular machine that can read messenger RNAs and translate the information they carry into proteins. There are many RNAs with other roles – in particular regulating which genes are expressed, but also as the genomes of most viruses. Ribose Nucleic Acids Most cellular RNA is single stranded, although some viruses have double stranded RNA. The single RNA strand is folded upon itself, either entirely or in certain regions. -

The Evolution of the Genetic Code: Impasses and Challenges
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Repository of the Academy's Library Special Issue on Code Biology in BioSystems The evolution of the genetic code: impasses and challenges Ádám Kun1,2,3 & Ádám Radványi4 1 Parmenides Center for the Conceptual Foundations of Science, Munich/Pullach, Germany. 2 MTA-ELTE Theoretical Biology and Evolutionary Ecology Research Group, Budapest, Hungary. 3 Evolutionary Systems Research Group, Centre for Ecological Research, Tihany, Hungary 4 Department of Plant Systematics, Ecology and Theoretical Biology, Institute of Biology, Eötvös University, Pázmány Péter sétány 1/C, 1117 Budapest, Hungary Abstract The origin of the genetic code and translation is a “notoriously difficult problem”. In this survey we present a list of questions that a full theory of the genetic code needs to answer. We assess the leading hypotheses according to these criteria. The stereochemical, the coding coenzyme handle, the coevolution, the four-column theory, the error minimization and the frozen accident hypotheses are discussed. The integration of these hypotheses can account for the origin of the genetic code. But experiments are badly needed. Thus we suggest a host of experiments that could (in)validate some of the models. We focus especially on the coding coenzyme handle hypothesis (CCH). The CCH suggests that amino acids attached to RNA handles enhanced catalytic activities of ribozymes. Alternatively, amino acids without handles or with a handle consisting of a single adenine, like in contemporary coenzymes could have been employed. All three scenarios can be tested in in vitro compartmentalized systems. Keywords Origin of Life; genetic code; RNA world; ribozyme; coding coenzyme handle; Introduction Modern cells store information in DNA and have peptide enzymes to carry out the metabolism for the cell.