Mrna Vaccine Era—Mechanisms, Drug Platform and Clinical Prospection
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Xfect™ RNA Transfection Reagent Protocol-At-A-Glance
Xfect™ RNA Transfection Reagent Protocol-At-A-Glance I. Introduction A. Summary This Protocol-At-A-Glance is provided for transfection of cells with RNA using the Xfect RNA Transfection Reagent (Cat. No. 631450). It describes transfection of mammalian cells with RNA (mRNA, sgRNA, microRNA, or shRNA) in a 12-well plate format. For formats other than 12-well plates, see Tables I and II for appropriate reaction volumes. Transfections can be carried out entirely in the presence of serum. The protocol is divided into two sections: Section II describes transfection of cells with RNA only, without the cotransfection of DNA. Section III describes cotransfection of cells with both RNA and DNA. NOTE: When transfecting cells with mRNA, expression may be increased by using serum-free medium during transfection, since serum may contain RNases that can reduce the amount of full-length mRNA. B. General Considerations Storage & handling Store the Xfect RNA Transfection Polymer at –20°C. Do not thaw until ready to use. Once thawed, store at 4°C for up to 12 months. NOTE: The Xfect RNA Transfection Polymer is a milky suspension and should be vortexed briefly prior to use to ensure that it is fully resuspended. Thaw Xfect Reaction Buffer at room temperature just prior to use. Vortex after thawing. Once thawed, store Xfect Reaction Buffer at 4°C for up to 12 months. Xfect Polymer The protocol for cotransfection with DNA (Section III) requires the use of the Xfect Polymer (not included; sold as part of the DNA Xfect Transfection Reagent, Cat. Nos. 631317, 631318). -

Human Plasma and Recombinant Hemopexins: Heme Binding Revisited
International Journal of Molecular Sciences Article Human Plasma and Recombinant Hemopexins: Heme Binding Revisited Elena Karnaukhova 1,*, Catherine Owczarek 2, Peter Schmidt 2, Dominik J. Schaer 3 and Paul W. Buehler 4,5,* 1 Center for Biologics Evaluation and Research, Food and Drug Administration, Silver Spring, MD 20993, USA 2 CSL Limited, Bio21 Institute, Parkville, Victoria 3010, Australia; [email protected] (C.O.); [email protected] (P.S.) 3 Division of Internal Medicine, University Hospital of Zurich, 8091 Zurich, Switzerland; [email protected] 4 Department of Pathology, The University of Maryland School of Medicine, Baltimore, MD 21201, USA 5 The Center for Blood Oxygen Transport and Hemostasis, Department of Pediatrics, The University of Maryland School of Medicine, Baltimore, MD 21201, USA * Correspondence: [email protected] (E.K.); [email protected] (P.W.B.) Abstract: Plasma hemopexin (HPX) is the key antioxidant protein of the endogenous clearance pathway that limits the deleterious effects of heme released from hemoglobin and myoglobin (the term “heme” is used in this article to denote both the ferrous and ferric forms). During intra-vascular hemolysis, heme partitioning to protein and lipid increases as the plasma concentration of HPX declines. Therefore, the development of HPX as a replacement therapy during high heme stress could be a relevant intervention for hemolytic disorders. A logical approach to enhance HPX yield involves recombinant production strategies from human cell lines. The present study focuses on a biophysical assessment of heme binding to recombinant human HPX (rhHPX) produced in the Expi293FTM (HEK293) cell system. -

Split Deoxyribozyme Probe for Efficient Detection of Highly Structured RNA Targets
University of Central Florida STARS Honors Undergraduate Theses UCF Theses and Dissertations 2018 Split Deoxyribozyme Probe For Efficient Detection of Highly Structured RNA Targets Sheila Raquel Solarez University of Central Florida Part of the Biochemistry Commons, and the Biology Commons Find similar works at: https://stars.library.ucf.edu/honorstheses University of Central Florida Libraries http://library.ucf.edu This Open Access is brought to you for free and open access by the UCF Theses and Dissertations at STARS. It has been accepted for inclusion in Honors Undergraduate Theses by an authorized administrator of STARS. For more information, please contact [email protected]. Recommended Citation Solarez, Sheila Raquel, "Split Deoxyribozyme Probe For Efficient Detection of Highly Structured RNA Targets" (2018). Honors Undergraduate Theses. 311. https://stars.library.ucf.edu/honorstheses/311 SPLIT DEOXYRIBOZYME PROBE FOR EFFICIENT DETECTION OF HIGHLY STRUCTURED RNA TARGETS By SHEILA SOLAREZ A thesis submitted in partial fulfillment of the requirements for the Honors in the Major Program in Biological Sciences in the College of Sciences and the Burnett Honors College at the University of Central Florida Orlando, Florida Spring Term, 2018 Thesis Chair: Yulia Gerasimova, PhD ABSTRACT Transfer RNAs (tRNAs) are known for their role as adaptors during translation of the genetic information and as regulators for gene expression; uncharged tRNAs regulate global gene expression in response to changes in amino acid pools in the cell. Aminoacylated tRNAs play a role in non-ribosomal peptide bond formation, post-translational protein labeling, modification of phospholipids in the cell membrane, and antibiotic biosynthesis. [1] tRNAs have a highly stable structure that can present a challenge for their detection using conventional techniques. -

Yeast Genome Gazetteer P35-65
gazetteer Metabolism 35 tRNA modification mitochondrial transport amino-acid metabolism other tRNA-transcription activities vesicular transport (Golgi network, etc.) nitrogen and sulphur metabolism mRNA synthesis peroxisomal transport nucleotide metabolism mRNA processing (splicing) vacuolar transport phosphate metabolism mRNA processing (5’-end, 3’-end processing extracellular transport carbohydrate metabolism and mRNA degradation) cellular import lipid, fatty-acid and sterol metabolism other mRNA-transcription activities other intracellular-transport activities biosynthesis of vitamins, cofactors and RNA transport prosthetic groups other transcription activities Cellular organization and biogenesis 54 ionic homeostasis organization and biogenesis of cell wall and Protein synthesis 48 plasma membrane Energy 40 ribosomal proteins organization and biogenesis of glycolysis translation (initiation,elongation and cytoskeleton gluconeogenesis termination) organization and biogenesis of endoplasmic pentose-phosphate pathway translational control reticulum and Golgi tricarboxylic-acid pathway tRNA synthetases organization and biogenesis of chromosome respiration other protein-synthesis activities structure fermentation mitochondrial organization and biogenesis metabolism of energy reserves (glycogen Protein destination 49 peroxisomal organization and biogenesis and trehalose) protein folding and stabilization endosomal organization and biogenesis other energy-generation activities protein targeting, sorting and translocation vacuolar and lysosomal -

Expanding the Genetic Code Lei Wang and Peter G
Reviews P. G. Schultz and L. Wang Protein Science Expanding the Genetic Code Lei Wang and Peter G. Schultz* Keywords: amino acids · genetic code · protein chemistry Angewandte Chemie 34 2005 Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim DOI: 10.1002/anie.200460627 Angew. Chem. Int. Ed. 2005, 44,34–66 Angewandte Protein Science Chemie Although chemists can synthesize virtually any small organic molecule, our From the Contents ability to rationally manipulate the structures of proteins is quite limited, despite their involvement in virtually every life process. For most proteins, 1. Introduction 35 modifications are largely restricted to substitutions among the common 20 2. Chemical Approaches 35 amino acids. Herein we describe recent advances that make it possible to add new building blocks to the genetic codes of both prokaryotic and 3. In Vitro Biosynthetic eukaryotic organisms. Over 30 novel amino acids have been genetically Approaches to Protein encoded in response to unique triplet and quadruplet codons including Mutagenesis 39 fluorescent, photoreactive, and redox-active amino acids, glycosylated 4. In Vivo Protein amino acids, and amino acids with keto, azido, acetylenic, and heavy-atom- Mutagenesis 43 containing side chains. By removing the limitations imposed by the existing 20 amino acid code, it should be possible to generate proteins and perhaps 5. An Expanded Code 46 entire organisms with new or enhanced properties. 6. Outlook 61 1. Introduction The genetic codes of all known organisms specify the same functional roles to amino acid residues in proteins. Selectivity 20 amino acid building blocks. These building blocks contain a depends on the number and reactivity (dependent on both limited number of functional groups including carboxylic steric and electronic factors) of a particular amino acid side acids and amides, a thiol and thiol ether, alcohols, basic chain. -

Nano-Lactoferrin in Diagnostic, Imaging and Targeted Delivery for Cancer and Infectious Diseases Jagat R
cer Scien an ce C & f o T Kanwar et al., J Cancer Sci Ther 2012, 4.3 l h a e n r a Journal of 1000107 r p u DOI: 10.4172/1948-5956. y o J ISSN: 1948-5956 Cancer Science & Therapy Review Article Open Access Nano-Lactoferrin in Diagnostic, Imaging and Targeted Delivery for Cancer and Infectious Diseases Jagat R. Kanwar1*, Rasika M. Samarasinghe1, Rakesh Sehgal2 and Rupinder K. Kanwar1 1Nanomedicine-Laboratory of Immunology and Molecular Biomedical Research (LIMBR), Centre for Biotechnology and Interdisciplinary Biosciences (BioDeakin), Institute for Technology & Research Innovation (ITRI), Deakin University, Geelong, Technology Precinct (GTP), Pigdons Road, Waurn Ponds, Geelong, Victoria 3217, Australia 2Professor, Department of Parasitology, Postgraduate Institute of Medical Education & Research, Chandigarh, India-160012 Abstract Lactoferrin (Lf) is a natural occurring iron binding protein present in many mammalian excretions and involved in various physiological processes. Lf is used in the transport of iron along with other molecules and ions from the digestive system. However its the modulatory functions exhibited by Lf in connection to immune response, disease regression and diagnosis that has made this protein an attractive therapeutic against chronic diseases. Further, the exciting potentials of employing nanotechnology in advancing drug delivery systems, active disease targeting and prognosis have also shown some encouraging outcomes. This review focuses on the role of Lf in diagnosing infection, cancer, neurological and inflammatory diseases and the recent nanotechnology based strategies. Keywords: Lactoferrin; Nanodelivery; Cancer; Inflammation; fluids, mucosal secretions, pancreatic fluids and in neutrophils cells [8]. Infection; Oral administration Structurally, Lf weighs approximately 80kDa and the polypeptide folds into two globular lobes. -

Genome-Scale Fitness Profile of Caulobacter Crescentus Grown in Natural Freshwater
Supplemental Material Genome-scale fitness profile of Caulobacter crescentus grown in natural freshwater Kristy L. Hentchel, Leila M. Reyes Ruiz, Aretha Fiebig, Patrick D. Curtis, Maureen L. Coleman, Sean Crosson Tn5 and Tn-Himar: comparing gene essentiality and the effects of gene disruption on fitness across studies A previous analysis of a highly saturated Caulobacter Tn5 transposon library revealed a set of genes that are required for growth in complex PYE medium [1]; approximately 14% of genes in the genome were deemed essential. The total genome insertion coverage was lower in the Himar library described here than in the Tn5 dataset of Christen et al (2011), as Tn-Himar inserts specifically into TA dinucleotide sites (with 67% GC content, TA sites are relatively limited in the Caulobacter genome). Genes for which we failed to detect Tn-Himar insertions (Table S13) were largely consistent with essential genes reported by Christen et al [1], with exceptions likely due to differential coverage of Tn5 versus Tn-Himar mutagenesis and differences in metrics used to define essentiality. A comparison of the essential genes defined by Christen et al and by our Tn5-seq and Tn-Himar fitness studies is presented in Table S4. We have uncovered evidence for gene disruptions that both enhanced or reduced strain fitness in lake water and M2X relative to PYE. Such results are consistent for a number of genes across both the Tn5 and Tn-Himar datasets. Disruption of genes encoding three metabolic enzymes, a class C β-lactamase family protein (CCNA_00255), transaldolase (CCNA_03729), and methylcrotonyl-CoA carboxylase (CCNA_02250), enhanced Caulobacter fitness in Lake Michigan water relative to PYE using both Tn5 and Tn-Himar approaches (Table S7). -

Differences in the Formation Mechanism of Giant Colonies in Two Phaeocystis Globosa Strains
International Journal of Molecular Sciences Article Differences in the Formation Mechanism of Giant Colonies in Two Phaeocystis globosa Strains 1,2, 2, 2 2, 1, Dayong Liang y, Xiaodong Wang y, Yiping Huo , Yan Wang * and Shaoshan Li * 1 Key Laboratory of Ecology and Environmental Science in Guangdong Higher Education, School of Life Science, South China Normal University, Guangzhou 510631, China; [email protected] 2 Research Center for Harmful Algae and Marine Biology, Jinan University, Guangzhou 510632, China; [email protected] (X.W.); [email protected] (Y.H.) * Correspondence: [email protected] (Y.W.); [email protected] (S.L.) The authors contributed equally to this work as co-first authors. y Received: 13 June 2020; Accepted: 27 July 2020; Published: 29 July 2020 Abstract: Phaeocystis globosa has become one of the primary causes of harmful algal bloom in coastal areas of southern China in recent years, and it poses a serious threat to the marine environment and other activities depending upon on it (e.g., aquaculture, cooling system of power plants), especially in the Beibu Gulf. We found colonies of P. globosa collected form Guangxi (China) were much larger than those obtained from Shantou cultured in lab. To better understand the causes of giant colonies formation, colonial cells collected from P. globosa GX strain (GX-C) and ST strain (ST-C) were separated by filtration. Morphological observations, phylogenetic analyses, rapid light-response curves, fatty acid profiling and transcriptome analyses of two type cells were performed in the laboratory. Although no differences in morphology and 18S rRNA sequences of these cells were observed, the colonies of GX strain (4.7 mm) are 30 times larger than those produced by the ST strain (300 µm). -

Small RNA Fragments Derived from Multiple RNA Classes – the Missing
Jackowiak et al. BMC Genomics (2017) 18:502 DOI 10.1186/s12864-017-3891-3 RESEARCHARTICLE Open Access Small RNA fragments derived from multiple RNA classes – the missing element of multi-omics characteristics of the hepatitis C virus cell culture model Paulina Jackowiak1, Anna Hojka-Osinska1, Anna Philips1, Agnieszka Zmienko1,2, Lucyna Budzko1, Patrick Maillard3, Agata Budkowska3,4 and Marek Figlerowicz1,2* Abstract Background: A pool of small RNA fragments (RFs) derived from diverse cellular RNAs has recently emerged as a rich source of functionally relevant molecules. Although their formation and accumulation has been connected to various stress conditions, the knowledge on RFs produced upon viral infections is very limited. Here, we applied the next generation sequencing (NGS) to characterize RFs generated in the hepatitis C virus (HCV) cell culture model (HCV-permissive Huh-7.5 cell line). Results: We found that both infected and non-infected cells contained a wide spectrum of RFs derived from virtually all RNA classes. A significant fraction of identified RFs accumulated to similar levels as miRNAs. Our analysis, focused on RFs originating from constitutively expressed non-coding RNAs, revealed three major patterns of parental RNA cleavage. We found that HCV infection induced significant changes in the accumulation of low copy number RFs, while subtly altered the levels of high copy number ones. Finally, the candidate RFs potentially relevant for host-virus interactions were identified. Conclusions: Our results indicate that RFs should be considered an important component of the Huh-7.5 transcriptome and suggest that the main factors influencing the RF biogenesis are the RNA structure and RNA protection by interacting proteins. -

Malaria Parasites Both Repress Host CXCL10 and Use It As a Cue for Growth Acceleration
ARTICLE https://doi.org/10.1038/s41467-021-24997-7 OPEN Malaria parasites both repress host CXCL10 and use it as a cue for growth acceleration Yifat Ofir-Birin 1, Hila Ben Ami Pilo1, Abel Cruz Camacho1, Ariel Rudik1, Anna Rivkin1, Or-Yam Revach1, Netta Nir1, Tal Block Tamin1, Paula Abou Karam1, Edo Kiper1, Yoav Peleg2, Reinat Nevo1, Aryeh Solomon3, Tal Havkin-Solomon1, Alicia Rojas1, Ron Rotkopf 4, Ziv Porat 5, Dror Avni6,7, Eli Schwartz6,7, Thomas Zillinger 8, Gunther Hartmann 8, Antonella Di Pizio 9, Neils Ben Quashie 10,11, Rivka Dikstein1, Motti Gerlic12, Ana Claudia Torrecilhas 13, Carmit Levy14, Esther N. M. Nolte-‘t Hoen15, ✉ Andrew G. Bowie 16 & Neta Regev-Rudzki 1 1234567890():,; Pathogens are thought to use host molecular cues to control when to initiate life-cycle transitions, but these signals are mostly unknown, particularly for the parasitic disease malaria caused by Plasmodium falciparum. The chemokine CXCL10 is present at high levels in fatal cases of cerebral malaria patients, but is reduced in patients who survive and do not have complications. Here we show a Pf ‘decision-sensing-system’ controlled by CXCL10 concentration. High CXCL10 expression prompts P. falciparum to initiate a survival strategy via growth acceleration. Remarkably, P. falciparum inhibits CXCL10 synthesis in monocytes by disrupting the association of host ribosomes with CXCL10 transcripts. The underlying inhi- bition cascade involves RNA cargo delivery into monocytes that triggers RIG-I, which leads to HUR1 binding to an AU-rich domain of the CXCL10 3’UTR. These data indicate that when the parasite can no longer keep CXCL10 at low levels, it can exploit the chemokine as a cue to shift tactics and escape. -
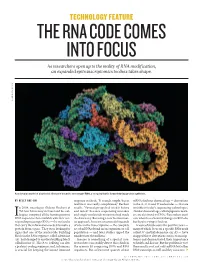
THE RNA CODE COMES INTO FOCUS As Researchers Open up to the Reality of RNA Modification, an Expanded Epitranscriptomics Toolbox Takes Shape
TECHNOLOGY FEATURE THE RNA CODE COMES INTO FOCUS As researchers open up to the reality of RNA modification, an expanded epitranscriptomics toolbox takes shape. LAGUNA DESIGN/SPL LAGUNA A molecular model of a bacterial ribosome bound to messenger RNA, a complex that is formed during protein synthesis. BY KELLY RAE CHI response in check. “It sounds simple, but in mRNAs harbour chemical tags — decorations real life it was really complicated,” Rechavi to the A, C, G and U nucleotides — that are n 2004, oncologist Gideon Rechavi at recalls. “Several groups had tried it before invisible to today’s sequencing technologies. Tel Aviv University in Israel and his col- and failed” because sequencing mistakes (Similar chemical tags, called epigenetic mark- leagues compared all the human genomic and single-nucleotide mutations had made ers, are also found on DNA.) Researchers aren’t IDNA sequences then available with their cor- the data noisy. But using a new bioinformat- sure what these chemical changes in RNA do, responding messenger RNAs — the molecules ics approach, his team uncovered thousands but they’re trying to find out. that carry the information needed to make a of sites in the transcriptome — the complete A wave of studies over the past five years — protein from a gene. They were looking for set of mRNAs found in an organism or cell many of which focus on a specific RNA mark signs that one of the nucleotide building population — and later studies upped the called N6-methyladenosine (m6A) — have blocks in the RNA sequence, called adenosine number into the millions1. -

The Role of the Salvage Pathway in Nucleotide Sugar Biosynthesis
THE ROLE OF THE SALVAGE PATHWAY IN NUCLEOTIDE SUGAR BIOSYNTHESIS: IDENTIFICATION OF SUGAR KINASES AND NDP-SUGAR PYROPHOSPHORYLASES by TING YANG (Under the Direction of Maor Bar-Peled) ABSTRACT The synthesis of polysaccharides, glycoproteins, glycolipids, glycosylated secondary metabolites and hormones requires a large number of glycosyltransferases and a constant supply of nucleotide sugars. In plants, photosynthesis and the NDP-sugar inter-conversion pathway are the major entry points to form NDP-sugars. In addition to these pathways is the salvage pathway, a less understood metabolism that provides the flux of NDP-sugars. This latter pathway involves the hydrolysis of glycans to free sugars, sugar transport, sugar phosphorylation and nucleotidylation. The balance between glycan synthesis and recycling as well as its regulation at various plant developmental stages remains elusive as many of the molecular components are unknown. To understand how the salvage pathway contributes to the sugar flux and cell wall biosynthesis, my research focused on the functional identification of salvage pathway sugar kinases and NDP-sugar pyrophosphorylases. This research led to the first identification and enzymatic characterization of galacturonic acid kinase (GalA kinase), galactokinase (GalK), a broad UDP-sugar pyrophosphorylase (sloppy), two promiscuous UDP-GlcNAc pyrophosphorylases (GlcNAc-1-P uridylyltransferases), as well as UDP-sugar pyrophosphorylase paralogs from Trypanosoma cruzi and Leishmania major. To evaluate the salvage pathway in plant biology, we further investigated a sugar kinase mutant: galacturonic acid kinase mutant (galak) and determined if and how galak KO mutant affects the synthesis of glycans in Arabidopsis. Feeding galacturonic acid to the seedlings exhibited a 40-fold accumulation of free GalA in galak mutant, while the wild type (WT) plant readily metabolizes the fed-sugar.