An Overview on the Development of Mrna-Based Vaccines and Their Formulation Strategies for Improved Antigen Expression in Vivo
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Rnai in Primary Cells and Difficult-To-Transfect Cell Lines
Automated High Throughput Nucleofection® RNAi in Primary Cells and Difficult-to-Transfect Cell Lines Claudia Merz, Bayer Schering Pharma AG, Berlin, Germany; Andreas Schroers, amaxa AG, Cologne, Germany; Eric Willimann, Tecan AG, Männedorf, Switzerland. Introduction Materials & Methods - Workflow Using primary cells for RNAi based applications such as target identification or – validation, requires a highly efficient transfection displaying the essential steps of the automated Nucleofector® Process: technology in combination with a reliable and robust automation system. To accomplish these requirements we integrated the amaxa 1. Transfer of the cells to the Nucleocuvette™ plate, 96-well Shuttle® in a Tecan Freedom EVO® cell transfection workstation which is based on Tecan’s Freedom EVO® liquid handling 2. Addition of the siRNA, (Steps 1 and 2 could be exchanged), platform and include all the necessary components and features for unattended cell transfection. 3. Nucleofection® process, 4. Addition of medium, Count Cells 5. Transfer of transfected cells to cell culture plate for incubation ® Nucleofector Technology prior to analysis. Remove Medium The 96-well Shuttle® combines high-throughput compatibility with the Nucleofector® Technology, which is a non-viral transfection method ideally suited for primary cells and hard-to-transfect cell lines based on a combination of buffers and electrical parameters. Nucleocuvette Plate Add Nucleofector +– The basic principle and benefits of the (empty) Solution Cell of interest Gene of interest Nucleofector® -

Main Outcomes of Discussion of WHO Consultation on Nucleic Acid
MAIN OUTCOMES OF DISCUSSION FROM WHO CONSULTATION ON NUCLEIC ACID VACCINES I. Knezevic, R. Sheets 21-23 Feb, 2018 Geneva, Switzerland CONTEXT OF DISCUSSION • WHO Consultation held to determine whether the existing DNA guidelines were due for revision or if they remained relevant with today’s status of nucleic acid vaccine development and maturity towards licensure (marketing authorization) • Presentation will cover alignment to existing guidelines • Current status of development of DNA and RNA vaccines, both prophylactic & therapeutic • Main outcomes of discussion • Next steps already underway & planned STATUS OF DEVELOPMENT OF NUCLEIC ACID VACCINES • First likely candidate to licensure could be a therapeutic DNA vaccine against Human Papilloma Viruses • Anticipated to be submitted for licensure in 3-5 years • Other DNA vaccines are likely to follow shortly thereafter, e.g., Zika prophylaxis • RNA vaccines – less clinical experience, but therapeutic RNAs anticipated in 2021/22 timeframe to be submitted for licensure • For priority pathogens in context of public health emergencies, several candidates under development (e.g., MERS-CoV, Marburg, Ebola) MAIN OUTCOMES - GENERAL • Regulators expressed need for updated DNA guideline for prophylaxis and therapy • Less need at present for RNA vaccines in guideline but need some basic PTC • Flexibility needed now until more experience gained for RNA vaccines that will come in next few years • Revisit need for a more specific guideline at appropriate time for RNA vaccines • Institutional Biosafety Committees are regulated by national jurisdictions & vary considerably • How can WHO assist to streamline or converge these review processes? Particularly, during Public Health Emergency – can anything be done beforehand? MAIN OUTCOMES - DEFINITIONS • Clear Definitions are needed • Proposed definition of DNA vaccine: • A DNA plasmid(s) into which the desired immunogen(s) is (are) encoded and prepared as purified plasmid preparations to be administered in vivo. -

Review of COVID-19 Vaccine Subtypes, Efficacy and Geographical Distributions Andre Ian Francis ,1 Saudah Ghany,1 Tia Gilkes,1 Srikanth Umakanthan2
Review Postgrad Med J: first published as 10.1136/postgradmedj-2021-140654 on 6 August 2021. Downloaded from Review of COVID-19 vaccine subtypes, efficacy and geographical distributions Andre Ian Francis ,1 Saudah Ghany,1 Tia Gilkes,1 Srikanth Umakanthan2 1Department of Clinical Medical ABSTRACT 2021, the Ad26.COV2.S, developed by Janssen Sciences, The Faculty of Medical As of 1 May 2021, there have been 152 661 445 (Johnson & Johnson) and Moderna on 30 April.4 Sciences, The University of the COVAX, coordinated by WHO, Gavi: The West Indies, St. Augustine, Covid-19 cases with 3 202 256 deaths globally. Trinidad and Tobago This pandemic led to the race to discover a vaccine Vaccine Alliance, the Coalition for Epidemic 2Pathology unit, Department to achieve herd immunity and curtail the damaging Preparedness Innovations (CEPI), acts as a of Paraclinical Sciences, The effects of Covid-19. This study aims to discuss the most programme that supports the development of University Of The West Indies, St. COVID-19 vaccine candidates and negotiates their Augustine, Trinidad and Tobago recent WHO-approved Covid-19 vaccine subtypes, their status and geographical scheduled updates as of 4 pricing to ensure low- and- middle- income countries have a fair shot at receiving vaccines.5 Correspondence to May 2021. The keywords “Covid-19, Vaccines, Pfizer, Mr. Andre Ian Francis, The BNT162b2, AstraZeneca, AZD1222, Moderna, mRNA- This article aims at discussing the most recent University of the West Indies, St. 1273, Janssen, Ad26.COV2.S” were typed into PubMed. WHO- approved COVID-19 vaccine subtypes, their Augustine, Trinidad and Tobago; Thirty Two relevant PubMed articles were included in status and geographical scheduled updates as of 4 andre. -

Split Deoxyribozyme Probe for Efficient Detection of Highly Structured RNA Targets
University of Central Florida STARS Honors Undergraduate Theses UCF Theses and Dissertations 2018 Split Deoxyribozyme Probe For Efficient Detection of Highly Structured RNA Targets Sheila Raquel Solarez University of Central Florida Part of the Biochemistry Commons, and the Biology Commons Find similar works at: https://stars.library.ucf.edu/honorstheses University of Central Florida Libraries http://library.ucf.edu This Open Access is brought to you for free and open access by the UCF Theses and Dissertations at STARS. It has been accepted for inclusion in Honors Undergraduate Theses by an authorized administrator of STARS. For more information, please contact [email protected]. Recommended Citation Solarez, Sheila Raquel, "Split Deoxyribozyme Probe For Efficient Detection of Highly Structured RNA Targets" (2018). Honors Undergraduate Theses. 311. https://stars.library.ucf.edu/honorstheses/311 SPLIT DEOXYRIBOZYME PROBE FOR EFFICIENT DETECTION OF HIGHLY STRUCTURED RNA TARGETS By SHEILA SOLAREZ A thesis submitted in partial fulfillment of the requirements for the Honors in the Major Program in Biological Sciences in the College of Sciences and the Burnett Honors College at the University of Central Florida Orlando, Florida Spring Term, 2018 Thesis Chair: Yulia Gerasimova, PhD ABSTRACT Transfer RNAs (tRNAs) are known for their role as adaptors during translation of the genetic information and as regulators for gene expression; uncharged tRNAs regulate global gene expression in response to changes in amino acid pools in the cell. Aminoacylated tRNAs play a role in non-ribosomal peptide bond formation, post-translational protein labeling, modification of phospholipids in the cell membrane, and antibiotic biosynthesis. [1] tRNAs have a highly stable structure that can present a challenge for their detection using conventional techniques. -

V.5 3/18/2021 1 COVID-19 Vaccine FAQ Sheet
COVID-19 Vaccine FAQ Sheet (updated 3/18/2021) The AST has received queries from transplant professionals and the community regarding the COVID-19 vaccine. The following FAQ was developed to relay information on the current state of knowledge. This document is subject to change and will be updated frequently as new information or data becomes available. What kinds of vaccines are available or under development to prevent COVID-19? There are currently several vaccine candidates in use or under development. In the United States, the Government is supporting six separate vaccine candidates. Several other vaccines are also undergoing development outside of the United States government sponsorship and further information can be found here: NYTimes Coronavirus Vaccine Tracker: https://www.nytimes.com/interactive/2020/science/coronavirus-vaccine- tracker.html Washington Post Vaccine Tracker: https://www.washingtonpost.com/graphics/2020/health/covid-vaccine-update- coronavirus/ v.5 3/18/2021 1 The types of vaccines are as follows (March 1, 2021) 1: Table 1: Vaccines Under Development or Available Through EUA Vaccine Type Compound Name [Sponsor] Clinical Notes Trial Phase mRNA mRNA-1273 [Moderna] Phase 3 Emergency use in U.S., E.U., other countries Approved in Canada BNT162b2 (Comirnaty) [Pfizer] Phase 2/3 Emergency use in U.S., E.U., other countries Also approved in Canada and other countries Replication- AZD1222 (Covishield) Phase 2/3 Emergency use defective [AstraZeneca] in U.K., India, adenoviral other countries vector (not U.S.) JNJ-78326735/Ad26.COV2.S -

Mrna Vaccine Era—Mechanisms, Drug Platform and Clinical Prospection
International Journal of Molecular Sciences Review mRNA Vaccine Era—Mechanisms, Drug Platform and Clinical Prospection 1, 1, 2 1,3, Shuqin Xu y, Kunpeng Yang y, Rose Li and Lu Zhang * 1 State Key Laboratory of Genetic Engineering, Institute of Genetics, School of Life Science, Fudan University, Shanghai 200438, China; [email protected] (S.X.); [email protected] (K.Y.) 2 M.B.B.S., School of Basic Medical Sciences, Peking University Health Science Center, Beijing 100191, China; [email protected] 3 Shanghai Engineering Research Center of Industrial Microorganisms, Shanghai 200438, China * Correspondence: [email protected]; Tel.: +86-13524278762 These authors contributed equally to this work. y Received: 30 July 2020; Accepted: 30 August 2020; Published: 9 September 2020 Abstract: Messenger ribonucleic acid (mRNA)-based drugs, notably mRNA vaccines, have been widely proven as a promising treatment strategy in immune therapeutics. The extraordinary advantages associated with mRNA vaccines, including their high efficacy, a relatively low severity of side effects, and low attainment costs, have enabled them to become prevalent in pre-clinical and clinical trials against various infectious diseases and cancers. Recent technological advancements have alleviated some issues that hinder mRNA vaccine development, such as low efficiency that exist in both gene translation and in vivo deliveries. mRNA immunogenicity can also be greatly adjusted as a result of upgraded technologies. In this review, we have summarized details regarding the optimization of mRNA vaccines, and the underlying biological mechanisms of this form of vaccines. Applications of mRNA vaccines in some infectious diseases and cancers are introduced. It also includes our prospections for mRNA vaccine applications in diseases caused by bacterial pathogens, such as tuberculosis. -
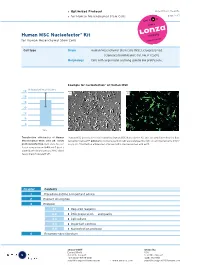
Optimized Protocol for Human Mesenchymal Stem Cells
› Optimized Protocol DPE-1001 Vs. 05-2005 › for Human Mesenchymal Stem Cells page 1 of 7 Human MSC Nucleofector® Kit for Human Mesenchymal Stem Cells Cell type Origin Human Mesenchymal Stem Cells (MSC), cryopreserved [Clonetics/BioWhittaker; Cat. No. PT-2501]. Morphology Cells with large nuclei and long spindle like protrusions.. Example for nucleofection® of Human MSC % transfection efficiency 60 A B 50 40 30 20 10 0 24 h Transfection efficiencies of Human Human MSC were nucleofected using the Human MSC Nucleofector Kit and a plasmid encoding the fluo- Mesenchymal Stem cells 24 hours rescent protein eGFP. 24 hours post-nucleofection cells were analyzed by light (A) and fluorescence micro- post nucleofection. Cells were nucleo- scopy (B). Transfection efficiencies of around 80% can be reached with eGFP. fected using program U-23 and 5 µg of a plasmid encoding the mouse MHC class I heavy chain molecule H-2Kk. Chapter Contents 1 Procedure outline & important advice 2 Product description 3 Protocol 3.1 › Required reagents 3.2 › DNA preparation and quality 3.3 › Cell culture 3.4 › Important controls 3.5 › Nucleofection protocol 4 Recommended literature 5 amaxa GmbH amaxa Inc. Europe/World USA Scientific Support Scientific Support +49 (0)221-99199-400 (240) 632-9110 [email protected] › www.amaxa.com [email protected] › Optimized Protocol DPE-1001 Vs. 05-2005 › for Human Mesenchymal Stem Cells page 2 of 7 1 Procedure outline & important advice Procedure outline Important advice 1. Preparation of cells. › Use MSCGM BulletKit (stored < 2 d at 4°C) (For details see 3.3.) › Passage interval: after reaching 70% confluency. -

Expanding the Genetic Code Lei Wang and Peter G
Reviews P. G. Schultz and L. Wang Protein Science Expanding the Genetic Code Lei Wang and Peter G. Schultz* Keywords: amino acids · genetic code · protein chemistry Angewandte Chemie 34 2005 Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim DOI: 10.1002/anie.200460627 Angew. Chem. Int. Ed. 2005, 44,34–66 Angewandte Protein Science Chemie Although chemists can synthesize virtually any small organic molecule, our From the Contents ability to rationally manipulate the structures of proteins is quite limited, despite their involvement in virtually every life process. For most proteins, 1. Introduction 35 modifications are largely restricted to substitutions among the common 20 2. Chemical Approaches 35 amino acids. Herein we describe recent advances that make it possible to add new building blocks to the genetic codes of both prokaryotic and 3. In Vitro Biosynthetic eukaryotic organisms. Over 30 novel amino acids have been genetically Approaches to Protein encoded in response to unique triplet and quadruplet codons including Mutagenesis 39 fluorescent, photoreactive, and redox-active amino acids, glycosylated 4. In Vivo Protein amino acids, and amino acids with keto, azido, acetylenic, and heavy-atom- Mutagenesis 43 containing side chains. By removing the limitations imposed by the existing 20 amino acid code, it should be possible to generate proteins and perhaps 5. An Expanded Code 46 entire organisms with new or enhanced properties. 6. Outlook 61 1. Introduction The genetic codes of all known organisms specify the same functional roles to amino acid residues in proteins. Selectivity 20 amino acid building blocks. These building blocks contain a depends on the number and reactivity (dependent on both limited number of functional groups including carboxylic steric and electronic factors) of a particular amino acid side acids and amides, a thiol and thiol ether, alcohols, basic chain. -

An Update on Self-Amplifying Mrna Vaccine Development
Review An Update on Self-Amplifying mRNA Vaccine Development Anna K. Blakney 1,* , Shell Ip 2 and Andrew J. Geall 2 1 Michael Smith Laboratories, School of Biomedical Engineering, University of British Columbia, Vancouver, BC V6T 1Z4, Canada 2 Precision NanoSystems Inc., Vancouver, BC V6P 6T7, Canada; [email protected] (S.I.); [email protected] (A.J.G.) * Correspondence: [email protected] Abstract: This review will explore the four major pillars required for design and development of an saRNA vaccine: Antigen design, vector design, non-viral delivery systems, and manufacturing (both saRNA and lipid nanoparticles (LNP)). We report on the major innovations, preclinical and clinical data reported in the last five years and will discuss future prospects. Keywords: RNA; self-amplifying RNA; replicon; vaccine; drug delivery 1. Introduction: The Four Pillars of saRNA Vaccines In December 2019, the SARS-CoV-2 (severe acute respiratory syndrome coronavirus 2) virus emerged, causing a respiratory illness, coronavirus disease 2019 (COVID-19), in Hubei province, China [1,2]. The virus has spread globally, with the World Health Organization (WHO) declaring it a Public Health Emergency of International concern on 30 January 2020 and a pandemic officially on 7 March 2020 [3]. There is a strong consensus globally that a COVID-19 vaccine is likely the most effective approach to sustainably controlling the COVID-19 pandemic [4]. There has been an unprecedented research effort and global Citation: Blakney, A.K.; Ip, S.; Geall, coordination which has resulted in the rapid development of vaccine candidates and A.J. An Update on Self-Amplifying initiation of human clinical trials. -

Small RNA Fragments Derived from Multiple RNA Classes – the Missing
Jackowiak et al. BMC Genomics (2017) 18:502 DOI 10.1186/s12864-017-3891-3 RESEARCHARTICLE Open Access Small RNA fragments derived from multiple RNA classes – the missing element of multi-omics characteristics of the hepatitis C virus cell culture model Paulina Jackowiak1, Anna Hojka-Osinska1, Anna Philips1, Agnieszka Zmienko1,2, Lucyna Budzko1, Patrick Maillard3, Agata Budkowska3,4 and Marek Figlerowicz1,2* Abstract Background: A pool of small RNA fragments (RFs) derived from diverse cellular RNAs has recently emerged as a rich source of functionally relevant molecules. Although their formation and accumulation has been connected to various stress conditions, the knowledge on RFs produced upon viral infections is very limited. Here, we applied the next generation sequencing (NGS) to characterize RFs generated in the hepatitis C virus (HCV) cell culture model (HCV-permissive Huh-7.5 cell line). Results: We found that both infected and non-infected cells contained a wide spectrum of RFs derived from virtually all RNA classes. A significant fraction of identified RFs accumulated to similar levels as miRNAs. Our analysis, focused on RFs originating from constitutively expressed non-coding RNAs, revealed three major patterns of parental RNA cleavage. We found that HCV infection induced significant changes in the accumulation of low copy number RFs, while subtly altered the levels of high copy number ones. Finally, the candidate RFs potentially relevant for host-virus interactions were identified. Conclusions: Our results indicate that RFs should be considered an important component of the Huh-7.5 transcriptome and suggest that the main factors influencing the RF biogenesis are the RNA structure and RNA protection by interacting proteins. -
THE RNA CODE COMES INTO FOCUS As Researchers Open up to the Reality of RNA Modification, an Expanded Epitranscriptomics Toolbox Takes Shape
TECHNOLOGY FEATURE THE RNA CODE COMES INTO FOCUS As researchers open up to the reality of RNA modification, an expanded epitranscriptomics toolbox takes shape. LAGUNA DESIGN/SPL LAGUNA A molecular model of a bacterial ribosome bound to messenger RNA, a complex that is formed during protein synthesis. BY KELLY RAE CHI response in check. “It sounds simple, but in mRNAs harbour chemical tags — decorations real life it was really complicated,” Rechavi to the A, C, G and U nucleotides — that are n 2004, oncologist Gideon Rechavi at recalls. “Several groups had tried it before invisible to today’s sequencing technologies. Tel Aviv University in Israel and his col- and failed” because sequencing mistakes (Similar chemical tags, called epigenetic mark- leagues compared all the human genomic and single-nucleotide mutations had made ers, are also found on DNA.) Researchers aren’t IDNA sequences then available with their cor- the data noisy. But using a new bioinformat- sure what these chemical changes in RNA do, responding messenger RNAs — the molecules ics approach, his team uncovered thousands but they’re trying to find out. that carry the information needed to make a of sites in the transcriptome — the complete A wave of studies over the past five years — protein from a gene. They were looking for set of mRNAs found in an organism or cell many of which focus on a specific RNA mark signs that one of the nucleotide building population — and later studies upped the called N6-methyladenosine (m6A) — have blocks in the RNA sequence, called adenosine number into the millions1. -

Methods of Transfection with Messenger RNA Gene Vectors
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Spiral - Imperial College Digital Repository Chapter 2 Methods of Transfection with Messenger RNA Gene Vectors Oleg E. Tolmachov and Tanya Tolmachova Additional information is available at the end of the chapter http://dx.doi.org/10.5772/61688 Abstract Non-viral gene delivery vectors with messenger RNA (mRNA) as a carrier of genetic information are among the staple gene transfer vectors for research in gene therapy, gene vaccination and cell fate reprogramming. As no passage of genetic cargo in and out of the nucleus is required, mRNA-based vectors typically offer the following five advantages: 1) fast start of transgene expression; 2) ability to express genes in non- dividing cells with an intact nuclear envelope; 3) insensitivity to the major gene silencing mechanisms, which operate in the nucleus; 4) absence of potentially mutagenic genomic insertions; 5) high cell survival rate after transfection procedures, which do not need to disturb nuclear envelope. In addition, mRNA-based vectors offer a simple combination of various transgenes through mixing of several mRNAs in a single multi-gene cocktail or expression of a number of proteins from a single mRNA molecule using internal ribosome entry sites (IRESes), ribosome skipping sequences and proteolytic signals. However, on the downside, uncontrolled extrac‐ ellular and intracellular decay of mRNA can be a substantial hurdle for mRNA- mediated gene transfer. Procedures for mRNA delivery are analogous to DNA transfer methods, which are well-established. In general, there are three actors in the gene delivery play, namely, the vector, the cell and the transfer environment.