Supplemental Tables
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Peripheral T Cells Ets-1 Maintains IL-7 Receptor Expression In
The Journal of Immunology Ets-1 Maintains IL-7 Receptor Expression in Peripheral T Cells Roland Grenningloh,*,† Tzong-Shyuan Tai,* Nicole Frahm,†,‡,1 Tomoyuki C. Hongo,‡ Adam T. Chicoine,‡ Christian Brander,†,‡,x,{ Daniel E. Kaufmann,†,‡,‖ and I-Cheng Ho*,† The expression of CD127, the IL-7–binding subunit of the IL-7 R, is tightly regulated during the development and activation of T cells and is reduced during chronic viral infection. However, the molecular mechanism regulating the dynamic expression of CD127 is still poorly understood. In this study, we report that the transcription factor Ets-1 is required for maintaining the expression of CD127 in murine peripheral T cells. Ets-1 binds to and activates the CD127 promoter, and its absence leads to reduced CD127 expression, attenuated IL-7 signaling, and impaired IL-7–dependent homeostatic proliferation of T cells. The expression of CD127 and Ets-1 is strongly correlated in human T cells. Both CD127 and Ets-1 expression are decreased in CD8+ T cells during HIV infection. In addition, HIV-associated loss of CD127 is only observed in Ets-1low effector memory and central memory but not in Ets-1high naive CD8+ T cells. Taken together, our data identify Ets-1 as a critical regulator of CD127 expression in T cells. The Journal of Immunology, 2011, 186: 969–976. nterleukin-7 signals are required for T cell development, GABPa or another Ets protein is responsible for maintaining maintaining the naive T cell pool, mounting proper primary CD127 expression in peripheral T cells is unknown. I responses, and inducing and maintaining CD4+ and CD8+ Ets-1 (E26 transformation-specific sequence) is the founding T cell memory (1–3). -
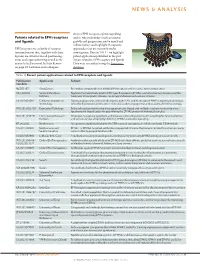
Patents Related to EPH Receptors and Ligands
NEWS & ANALYSIS discuss EPH receptor–ephrin signalling Patents related to EPH receptors and its role in disorders such as tumour and ligands growth and progression, nerve injury and inflammation, and highlight therapeutic EPH receptors are a family of receptor approaches that are currently under tyrosine kinases that, together with their investigation. Here in TABLE 1 we highlight ligands, are involved in cell positioning, patent applications published in the past tissue and organ patterning as well as the 3 years related to EPH receptors and ligands. control of cell survival. In their Review Data were researched using the Espacenet on page 39, Lackman and colleagues database. Table 1 | Recent patent applications related to EPH receptors and ligands Nature Reviews | Drug Discovery Publication Applicants Subject numbers NZ 581397 AstraZeneca Pyrimidine compounds that inhibit EPH receptors and are useful for treating cancer HK 1108702 Sanford-Burnham Peptides that selectively bind to EPH type-B receptors (EPHBs); useful for tumour imaging and the Institute treatment of neoplastic disease, neurological disease and vascular disease US 2013091591 California Institute of During angiogenesis, arterial cells express ephrin B2, and its receptor EPHB4 is expressed on venous Technology cells; this distinction can be used in methods to alter angiogenesis and to assess the effect of drugs WO 2013052710 Expression Pathology Selected reaction monitoring mass spectrometry-based and multiple reaction monitoring mass spectrometry-based assays for quantifying -

Cyclin D1 Is a Direct Transcriptional Target of GATA3 in Neuroblastoma Tumor Cells
Oncogene (2010) 29, 2739–2745 & 2010 Macmillan Publishers Limited All rights reserved 0950-9232/10 $32.00 www.nature.com/onc SHORT COMMUNICATION Cyclin D1 is a direct transcriptional target of GATA3 in neuroblastoma tumor cells JJ Molenaar1,2, ME Ebus1, J Koster1, E Santo1, D Geerts1, R Versteeg1 and HN Caron2 1Department of Human Genetics, Academic Medical Center, University of Amsterdam, Amsterdam, The Netherlands and 2Department of Pediatric Oncology, Emma Kinderziekenhuis, Academic Medical Center, University of Amsterdam, Amsterdam, The Netherlands Almost all neuroblastoma tumors express excess levels of 2000). Several checkpoints normally prevent premature Cyclin D1 (CCND1) compared to normal tissues and cell-cycle progression and cell division. The crucial G1 other tumor types. Only a small percentage of these entry point is controlled by the D-type Cyclins that can neuroblastoma tumors have high-level amplification of the activate CDK4/6 that in turn phosphorylate the pRb Cyclin D1 gene. The other neuroblastoma tumors have protein. This results in a release of the E2F transcription equally high Cyclin D1 expression without amplification. factor that causes transcriptional upregulation of Silencing of Cyclin D1 expression was previously found to numerous genes involved in further progression of the trigger differentiation of neuroblastoma cells. Over- cell cycle (Sherr, 1996). expression of Cyclin D1 is therefore one of the most Neuroblastomas are embryonal tumors that originate frequent mechanisms with a postulated function in neuro- from precursor cells of the sympathetic nervous system. blastoma pathogenesis. The cause for the Cyclin D1 This tumor has a very poor prognosis and despite the overexpression is unknown. -

Characterization of BRCA1-Deficient Premalignant Tissues and Cancers Identifies Plekha5 As a Tumor Metastasis Suppressor
ARTICLE https://doi.org/10.1038/s41467-020-18637-9 OPEN Characterization of BRCA1-deficient premalignant tissues and cancers identifies Plekha5 as a tumor metastasis suppressor Jianlin Liu1,2, Ragini Adhav1,2, Kai Miao1,2, Sek Man Su1,2, Lihua Mo1,2, Un In Chan1,2, Xin Zhang1,2, Jun Xu1,2, Jianjie Li1,2, Xiaodong Shu1,2, Jianming Zeng 1,2, Xu Zhang1,2, Xueying Lyu1,2, Lakhansing Pardeshi1,3, ✉ ✉ Kaeling Tan1,3, Heng Sun1,2, Koon Ho Wong 1,3, Chuxia Deng 1,2 & Xiaoling Xu 1,2 1234567890():,; Single-cell whole-exome sequencing (scWES) is a powerful approach for deciphering intra- tumor heterogeneity and identifying cancer drivers. So far, however, simultaneous analysis of single nucleotide variants (SNVs) and copy number variations (CNVs) of a single cell has been challenging. By analyzing SNVs and CNVs simultaneously in bulk and single cells of premalignant tissues and tumors from mouse and human BRCA1-associated breast cancers, we discover an evolution process through which the tumors initiate from cells with SNVs affecting driver genes in the premalignant stage and malignantly progress later via CNVs acquired in chromosome regions with cancer driver genes. These events occur randomly and hit many putative cancer drivers besides p53 to generate unique genetic and pathological features for each tumor. Upon this, we finally identify a tumor metastasis suppressor Plekha5, whose deficiency promotes cancer metastasis to the liver and/or lung. 1 Cancer Centre, Faculty of Health Sciences, University of Macau, Macau, SAR, China. 2 Centre for Precision Medicine Research and Training, Faculty of Health Sciences, University of Macau, Macau, SAR, China. -

A Review of VEGF/VEGFR-Targeted Therapeutics for Recurrent Glioblastoma
414 Original Article A Review of VEGF/VEGFR-Targeted Therapeutics for Recurrent Glioblastoma David A. Reardon, MDa,b; Scott Turner, MDc; Katherine B. Peters, MD, PhDc; Annick Desjardins, MDc; Sridharan Gururangan, MDa,b; John H. Sampson, MD, PhDa; Roger E. McLendon, MDd; James E. Herndon II, PhDe; Lee W. Jones, PhDf; John P. Kirkpatrick, MD, PhDf; Allan H. Friedman, MDa; James J. Vredenburgh, MDc; Darell D. Bigner, MD, PhDd; and Henry S. Friedman, MDa,b; Durham, North Carolina Key Words ability effect of VEGF inhibitors, the Radiologic Assessment in Neu- Glioblastoma, angiogenesis, vascular endothelial growth factor, ro-Oncology (RANO) criteria were recently implemented to bet- malignant glioma ter assess response among patients with glioblastoma. Although bevacizumab improves survival and quality of life, eventual tumor progression is the norm. Better understanding of resistance mech- Abstract anisms to VEGF inhibitors and identification of effective therapy Glioblastoma, the most common primary malignant brain tumor after bevacizumab progression are currently a critical need for pa- among adults, is a highly angiogenic and deadly tumor. Angiogen- tients with glioblastoma. (JNCCN 2011;9:414–427) esis in glioblastoma, driven by hypoxia-dependent and indepen- dent mechanisms, is primarily mediated by vascular endothelial growth factor (VEGF), and generates blood vessels with distinctive features. The outcome for patients with recurrent glioblastoma is poor because of ineffective therapies. However, recent encourag- ing rates of radiographic response and progression-free survival, Malignant gliomas, including the most common sub- and adequate safety, led the FDA to grant accelerated approval of type of glioblastoma, are rapidly growing destructive tu- bevacizumab, a humanized monoclonal antibody against VEGF, for mors that extensively invade locally but rarely metasta- the treatment of recurrent glioblastoma in May 2009. -

Agonists and Knockdown of Estrogen Receptor Β Differentially Affect
Schüler-Toprak et al. BMC Cancer (2016) 16:951 DOI 10.1186/s12885-016-2973-y RESEARCH ARTICLE Open Access Agonists and knockdown of estrogen receptor β differentially affect invasion of triple-negative breast cancer cells in vitro Susanne Schüler-Toprak1*, Julia Häring1, Elisabeth C. Inwald1, Christoph Moehle2, Olaf Ortmann1 and Oliver Treeck1 Abstract Background: Estrogen receptor β (ERβ) is expressed in the majority of invasive breast cancer cases, irrespective of their subtype, including triple-negative breast cancer (TNBC). Thus, ERβ might be a potential target for therapy of this challenging cancer type. In this in vitro study, we examined the role of ERβ in invasion of two triple-negative breast cancer cell lines. Methods: MDA-MB-231 and HS578T breast cancer cells were treated with the specific ERβ agonists ERB-041, WAY200070, Liquiritigenin and 3β-Adiol. Knockdown of ERβ expression was performed by means of siRNA transfection. Effects on cellular invasion were assessed in vitro by means of a modified Boyden chamber assay. Transcriptome analyses were performed using Affymetrix Human Gene 1.0 ST microarrays. Pathway and gene network analyses were performed by means of Genomatix and Ingenuity Pathway Analysis software. Results: Invasiveness of MBA-MB-231 and HS578T breast cancer cells decreased after treatment with ERβ agonists ERB-041 and WAY200070. Agonists Liquiritigenin and 3β-Adiol only reduced invasion of MDA-MB-231 cells. Knockdown of ERβ expression increased invasiveness of MDA-MB-231 cells about 3-fold. Transcriptome and pathway analyses revealed that ERβ knockdown led to activation of TGFβ signalling and induced expression of a network of genes with functions in extracellular matrix, tumor cell invasion and vitamin D3 metabolism. -

Accompanies CD8 T Cell Effector Function Global DNA Methylation
Global DNA Methylation Remodeling Accompanies CD8 T Cell Effector Function Christopher D. Scharer, Benjamin G. Barwick, Benjamin A. Youngblood, Rafi Ahmed and Jeremy M. Boss This information is current as of October 1, 2021. J Immunol 2013; 191:3419-3429; Prepublished online 16 August 2013; doi: 10.4049/jimmunol.1301395 http://www.jimmunol.org/content/191/6/3419 Downloaded from Supplementary http://www.jimmunol.org/content/suppl/2013/08/20/jimmunol.130139 Material 5.DC1 References This article cites 81 articles, 25 of which you can access for free at: http://www.jimmunol.org/content/191/6/3419.full#ref-list-1 http://www.jimmunol.org/ Why The JI? Submit online. • Rapid Reviews! 30 days* from submission to initial decision • No Triage! Every submission reviewed by practicing scientists by guest on October 1, 2021 • Fast Publication! 4 weeks from acceptance to publication *average Subscription Information about subscribing to The Journal of Immunology is online at: http://jimmunol.org/subscription Permissions Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Email Alerts Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts The Journal of Immunology is published twice each month by The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2013 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. The Journal of Immunology Global DNA Methylation Remodeling Accompanies CD8 T Cell Effector Function Christopher D. Scharer,* Benjamin G. Barwick,* Benjamin A. Youngblood,*,† Rafi Ahmed,*,† and Jeremy M. -

Development and Validation of a Novel Immune-Related Prognostic Model
Wang et al. J Transl Med (2020) 18:67 https://doi.org/10.1186/s12967-020-02255-6 Journal of Translational Medicine RESEARCH Open Access Development and validation of a novel immune-related prognostic model in hepatocellular carcinoma Zheng Wang1, Jie Zhu1, Yongjuan Liu3, Changhong Liu2, Wenqi Wang2, Fengzhe Chen1* and Lixian Ma1* Abstract Background: Growing evidence has suggested that immune-related genes play crucial roles in the development and progression of hepatocellular carcinoma (HCC). Nevertheless, the utility of immune-related genes for evaluating the prognosis of HCC patients are still lacking. The study aimed to explore gene signatures and prognostic values of immune-related genes in HCC. Methods: We comprehensively integrated gene expression data acquired from 374 HCC and 50 normal tissues in The Cancer Genome Atlas (TCGA). Diferentially expressed genes (DEGs) analysis and univariate Cox regression analysis were performed to identify DEGs that related to overall survival. An immune prognostic model was constructed using the Lasso and multivariate Cox regression analyses. Furthermore, Cox regression analysis was applied to identify independent prognostic factors in HCC. The correlation analysis between immune-related signature and immune cells infltration were also investigated. Finally, the signature was validated in an external independent dataset. Results: A total of 329 diferentially expressed immune‐related genes were detected. 64 immune‐related genes were identifed to be markedly related to overall survival in HCC patients using univariate Cox regression analysis. Then we established a TF-mediated network for exploring the regulatory mechanisms of these genes. Lasso and multivariate Cox regression analyses were applied to construct the immune-based prognostic model, which consisted of nine immune‐related genes. -

A Tyrosine Kinase Expression Signature Predicts the Post-Operative Clinical Outcome in Triple Negative Breast Cancers
cancers Article A Tyrosine Kinase Expression Signature Predicts the Post-Operative Clinical Outcome in Triple Negative Breast Cancers Alexandre de Nonneville 1, Pascal Finetti 2 , José Adelaide 2, Éric Lambaudie 3, Patrice Viens 1, Anthony Gonçalves 1 , Daniel Birnbaum 2, Emilie Mamessier 2 and François Bertucci 1,2,* 1 Department of Medical Oncology, Institut Paoli-Calmettes, Aix-Marseille Univ, CRCM, CNRS, INSERM, 13000 Marseille, France 2 Laboratory of Predictive Oncology, Centre de Recherche en Cancérologie de Marseille, Institut Paoli-Calmettes, Inserm UMR1068, CNRS UMR725, Aix-Marseille Université, 13000 Marseille, France 3 Department of Surgical Oncology, Institut Paoli-Calmettes, Aix-Marseille Univ, CNRS, INSERM, CRCM, 13000 Marseille, France * Correspondence: [email protected]; Tel.: +33-4-91-22-35-37; Fax: +33-4-91-22-36-70 Received: 15 July 2019; Accepted: 9 August 2019; Published: 13 August 2019 Abstract: Triple negative breast cancer (TNBC) represent 15% of breast cancers. Histoclinical features and marketed prognostic gene expression signatures (GES) failed to identify good- and poor-prognosis patients. Tyrosine kinases (TK) represent potential prognostic and/or therapeutic targets for TNBC. We sought to define a prognostic TK GES in a large series of TNBC. mRNA expression and histoclinical data of 6379 early BCs were collected from 16 datasets. We searched for a TK-based GES associated with disease-free survival (DFS) and tested its robustness in an independent validation set. A total of 1226 samples were TNBC. In the learning set of samples (N = 825), we identified a 13-TK GES associated with DFS. This GES was associated with cell proliferation and immune response. -

Functional Analysis of Somatic Mutations Affecting Receptor Tyrosine Kinase Family in Metastatic Colorectal Cancer
Author Manuscript Published OnlineFirst on March 29, 2019; DOI: 10.1158/1535-7163.MCT-18-0582 Author manuscripts have been peer reviewed and accepted for publication but have not yet been edited. Functional analysis of somatic mutations affecting receptor tyrosine kinase family in metastatic colorectal cancer Leslie Duplaquet1, Martin Figeac2, Frédéric Leprêtre2, Charline Frandemiche3,4, Céline Villenet2, Shéhérazade Sebda2, Nasrin Sarafan-Vasseur5, Mélanie Bénozène1, Audrey Vinchent1, Gautier Goormachtigh1, Laurence Wicquart6, Nathalie Rousseau3, Ludivine Beaussire5, Stéphanie Truant7, Pierre Michel8, Jean-Christophe Sabourin9, Françoise Galateau-Sallé10, Marie-Christine Copin1,6, Gérard Zalcman11, Yvan De Launoit1, Véronique Fafeur1 and David Tulasne1 1 Univ. Lille, CNRS, Institut Pasteur de Lille, UMR 8161 - M3T – Mechanisms of Tumorigenesis and Target Therapies, F-59000 Lille, France. 2 Univ. Lille, Plateau de génomique fonctionnelle et structurale, CHU Lille, F-59000 Lille, France 3 TCBN - Tumorothèque Caen Basse-Normandie, F-14000 Caen, France. 4 Réseau Régional de Cancérologie – OncoBasseNormandie – F14000 Caen – France. 5 Normandie Univ, UNIROUEN, Inserm U1245, IRON group, Rouen University Hospital, Normandy Centre for Genomic and Personalized Medicine, F-76000 Rouen, France. 6 Tumorothèque du C2RC de Lille, F-59037 Lille, France. 7 Department of Digestive Surgery and Transplantation, CHU Lille, Univ Lille, 2 Avenue Oscar Lambret, 59037, Lille Cedex, France. 8 Department of hepato-gastroenterology, Rouen University Hospital, Normandie Univ, UNIROUEN, Inserm U1245, IRON group, F-76000 Rouen, France. 9 Department of Pathology, Normandy University, INSERM 1245, Rouen University Hospital, F 76 000 Rouen, France. 10 Department of Pathology, MESOPATH-MESOBANK, Centre León Bérard, Lyon, France. 11 Thoracic Oncology Department, CIC1425/CLIP2 Paris-Nord, Hôpital Bichat-Claude Bernard, Paris, France. -

Identifying Isl1 Genetic Lineage in the Developing Olfactory System and in Gnrh-1 Neurons 2 3 Ed Zandro M
bioRxiv preprint doi: https://doi.org/10.1101/2020.08.31.276360; this version posted August 31, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. 1 Identifying Isl1 genetic lineage in the developing olfactory system and in GnRH-1 neurons 2 3 Ed Zandro M. Taroc1, Raghu Ram Katreddi1 and Paolo E. Forni1*. 4 5 1Department of Biological Sciences; The RNA Institute, and the Center for Neuroscience 6 Research; University at Albany, State University of New York, Albany, NY 12222, USA. 7 8 * Corresponding Author 9 [email protected] 10 11 12 Keywords: Olfactory neurons, vomeronasal sensory neurons, GnRH neurons, Islet-1/Isl1, Isl1 13 Conditional knock-out; genetic lineage tracing, olfactory placode, neural crest. 14 15 Abstract (299, at 245 after editing) 16 During embryonic development, symmetric ectodermal thickenings (olfactory placodes) give rise 17 to several cell types that comprise the olfactory system, such as those that form the terminal nerve 18 ganglion (TN), gonadotropin releasing hormone-1 (GnRH-1) neurons and other migratory 19 neurons in rodents. Even though the genetic heterogeneity among these cell types are 20 documented, unidentified cell populations arising from the olfactory placode remain. One 21 candidate to identify placodal derived neurons in the developing nasal area is the transcription 22 factor Isl1, which was recently identified in GnRH-3 neurons of the terminal nerve in fish, as well 23 as expression in neurons of the nasal migratory mass. Here, we analyzed the Isl1 genetic lineage 24 in chemosensory neuronal populations in the nasal area and migratory GnRH-1 neurons in mice 25 using in-situ hybridization, immunolabeling a Tamoxifen inducible Isl1CreERT and a constitutive 26 Isl1Cre knock-in mouse lines. -

Ephrin-A Binding and Epha Receptor Expression Delineate the Matrix Compartment of the Striatum
The Journal of Neuroscience, June 15, 1999, 19(12):4962–4971 Ephrin-A Binding and EphA Receptor Expression Delineate the Matrix Compartment of the Striatum L. Scott Janis, Robert M. Cassidy, and Lawrence F. Kromer Department of Cell Biology and Interdisciplinary Program in Neuroscience, Georgetown University Medical Center, Washington, DC 20007 The striatum integrates limbic and neocortical inputs to regulate matrix neurons. In situ hybridization for EphA RTKs reveals that sensorimotor and psychomotor behaviors. This function is de- the two different ligand binding patterns strictly match pendent on the segregation of striatal projection neurons into the mRNA expression patterns of EphA4 and EphA7. anatomical and functional components, such as the striosome Ligand–receptor binding assays indicate that ephrin-A1 and and matrix compartments. In the present study the association ephrin-A4 selectively bind EphA4 but not EphA7 in the lysates of ephrin-A cell surface ligands and EphA receptor tyrosine of striatal tissue. Conversely, ephrin-A2, ephrin-A3, and kinases (RTKs) with the organization of these compartments ephrin-A5 bind EphA7 but not EphA4. These observations im- was determined in postnatal rats. Ephrin-A1 and ephrin-A4 plicate selective interactions between ephrin-A molecules and selectively bind to EphA receptors on neurons restricted to the EphA RTKs as potential mechanisms for regulating the com- matrix compartment. Binding is absent from the striosomes, partmental organization of the striatum. which were identified by m-opioid