Acquired Resistance to Dasatinib in Lung Cancer Cell Lines Conferred by DDR2 Gatekeeper Mutation and NF1 Loss
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
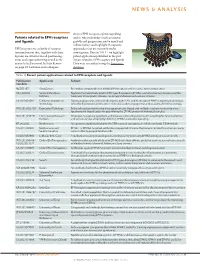
Patents Related to EPH Receptors and Ligands
NEWS & ANALYSIS discuss EPH receptor–ephrin signalling Patents related to EPH receptors and its role in disorders such as tumour and ligands growth and progression, nerve injury and inflammation, and highlight therapeutic EPH receptors are a family of receptor approaches that are currently under tyrosine kinases that, together with their investigation. Here in TABLE 1 we highlight ligands, are involved in cell positioning, patent applications published in the past tissue and organ patterning as well as the 3 years related to EPH receptors and ligands. control of cell survival. In their Review Data were researched using the Espacenet on page 39, Lackman and colleagues database. Table 1 | Recent patent applications related to EPH receptors and ligands Nature Reviews | Drug Discovery Publication Applicants Subject numbers NZ 581397 AstraZeneca Pyrimidine compounds that inhibit EPH receptors and are useful for treating cancer HK 1108702 Sanford-Burnham Peptides that selectively bind to EPH type-B receptors (EPHBs); useful for tumour imaging and the Institute treatment of neoplastic disease, neurological disease and vascular disease US 2013091591 California Institute of During angiogenesis, arterial cells express ephrin B2, and its receptor EPHB4 is expressed on venous Technology cells; this distinction can be used in methods to alter angiogenesis and to assess the effect of drugs WO 2013052710 Expression Pathology Selected reaction monitoring mass spectrometry-based and multiple reaction monitoring mass spectrometry-based assays for quantifying -

A Review of VEGF/VEGFR-Targeted Therapeutics for Recurrent Glioblastoma
414 Original Article A Review of VEGF/VEGFR-Targeted Therapeutics for Recurrent Glioblastoma David A. Reardon, MDa,b; Scott Turner, MDc; Katherine B. Peters, MD, PhDc; Annick Desjardins, MDc; Sridharan Gururangan, MDa,b; John H. Sampson, MD, PhDa; Roger E. McLendon, MDd; James E. Herndon II, PhDe; Lee W. Jones, PhDf; John P. Kirkpatrick, MD, PhDf; Allan H. Friedman, MDa; James J. Vredenburgh, MDc; Darell D. Bigner, MD, PhDd; and Henry S. Friedman, MDa,b; Durham, North Carolina Key Words ability effect of VEGF inhibitors, the Radiologic Assessment in Neu- Glioblastoma, angiogenesis, vascular endothelial growth factor, ro-Oncology (RANO) criteria were recently implemented to bet- malignant glioma ter assess response among patients with glioblastoma. Although bevacizumab improves survival and quality of life, eventual tumor progression is the norm. Better understanding of resistance mech- Abstract anisms to VEGF inhibitors and identification of effective therapy Glioblastoma, the most common primary malignant brain tumor after bevacizumab progression are currently a critical need for pa- among adults, is a highly angiogenic and deadly tumor. Angiogen- tients with glioblastoma. (JNCCN 2011;9:414–427) esis in glioblastoma, driven by hypoxia-dependent and indepen- dent mechanisms, is primarily mediated by vascular endothelial growth factor (VEGF), and generates blood vessels with distinctive features. The outcome for patients with recurrent glioblastoma is poor because of ineffective therapies. However, recent encourag- ing rates of radiographic response and progression-free survival, Malignant gliomas, including the most common sub- and adequate safety, led the FDA to grant accelerated approval of type of glioblastoma, are rapidly growing destructive tu- bevacizumab, a humanized monoclonal antibody against VEGF, for mors that extensively invade locally but rarely metasta- the treatment of recurrent glioblastoma in May 2009. -

Original Article ERBB3, IGF1R, and TGFBR2 Expression Correlate With
Am J Cancer Res 2018;8(5):792-809 www.ajcr.us /ISSN:2156-6976/ajcr0077452 Original Article ERBB3, IGF1R, and TGFBR2 expression correlate with PDGFR expression in glioblastoma and participate in PDGFR inhibitor resistance of glioblastoma cells Kang Song1,2*, Ye Yuan1,2*, Yong Lin1,2, Yan-Xia Wang1,2, Jie Zhou1,2, Qu-Jing Gai1,2, Lin Zhang1,2, Min Mao1,2, Xiao-Xue Yao1,2, Yan Qin1,2, Hui-Min Lu1,2, Xiang Zhang1,2, You-Hong Cui1,2, Xiu-Wu Bian1,2, Xia Zhang1,2, Yan Wang1,2 1Department of Pathology, Institute of Pathology and Southwest Cancer Center, Southwest Hospital, Third Military Medical University, Chongqing 400038, China; 2Key Laboratory of Tumor Immunology and Pathology of Ministry of Education, Chongqing 400038, China. *Equal contributors. Received April 6, 2018; Accepted April 9, 2018; Epub May 1, 2018; Published May 15, 2018 Abstract: Glioma, the most prevalent malignancy in brain, is classified into four grades (I, II, III, and IV), and grade IV glioma is also known as glioblastoma multiforme (GBM). Aberrant activation of receptor tyrosine kinases (RTKs), including platelet-derived growth factor receptor (PDGFR), are frequently observed in glioma. Accumulating evi- dence suggests that PDGFR plays critical roles during glioma development and progression and is a promising drug target for GBM therapy. However, PDGFR inhibitor (PDGFRi) has failed in clinical trials, at least partially, due to the activation of other RTKs, which compensates for PDGFR inhibition and renders tumor cells resistance to PDGFRi. Therefore, identifying the RTKs responsible for PDGFRi resistance might provide new therapeutic targets to syner- getically enhance the efficacy of PDGFRi. -

Erbb3 Is Involved in Activation of Phosphatidylinositol 3-Kinase by Epidermal Growth Factor STEPHEN P
MOLECULAR AND CELLULAR BIOLOGY, June 1994, p. 3550-3558 Vol. 14, No. 6 0270-7306/94/$04.00+0 Copyright C 1994, American Society for Microbiology ErbB3 Is Involved in Activation of Phosphatidylinositol 3-Kinase by Epidermal Growth Factor STEPHEN P. SOLTOFF,l* KERMIT L. CARRAWAY III,1 S. A. PRIGENT,2 W. G. GULLICK,2 AND LEWIS C. CANTLEY' Division of Signal Transduction, Department ofMedicine, Beth Israel Hospital, Boston, Massachusetts 02115,1 and Molecular Oncology Laboratory, ICRF Oncology Group, Hammersmith Hospital, London W12 OHS, United Kingdom2 Received 11 October 1993/Returned for modification 11 November 1993/Accepted 24 February 1994 Conflicting results concerning the ability of the epidermal growth factor (EGF) receptor to associate with and/or activate phosphatidylinositol (Ptdlns) 3-kinase have been published. Despite the ability of EGF to stimulate the production of Ptdlns 3-kinase products and to cause the appearance of PtdIns 3-kinase activity in antiphosphotyrosine immunoprecipitates in several cell lines, we did not detect EGF-stimulated Ptdlns 3-kinase activity in anti-EGF receptor immunoprecipitates. This result is consistent with the lack of a phosphorylated Tyr-X-X-Met motif, the p85 Src homology 2 (SH2) domain recognition sequence, in this receptor sequence. The EGF receptor homolog, ErbB2 protein, also lacks this motif. However, the ErbB3 protein has seven repeats of the Tyr-X-X-Met motif in the carboxy-terminal unique domain. Here we show that in A431 cells, which express both the EGF receptor and ErbB3, Ptdlns 3-kinase coprecipitates with the ErbB3 protein (pl80eR3) in response to EGF. p180B3 is also shown to be tyrosine phosphorylated in response to EGF. -

RET Gene Fusions in Malignancies of the Thyroid and Other Tissues
G C A T T A C G G C A T genes Review RET Gene Fusions in Malignancies of the Thyroid and Other Tissues Massimo Santoro 1,*, Marialuisa Moccia 1, Giorgia Federico 1 and Francesca Carlomagno 1,2 1 Department of Molecular Medicine and Medical Biotechnology, University of Naples “Federico II”, 80131 Naples, Italy; [email protected] (M.M.); [email protected] (G.F.); [email protected] (F.C.) 2 Institute of Endocrinology and Experimental Oncology of the CNR, 80131 Naples, Italy * Correspondence: [email protected] Received: 10 March 2020; Accepted: 12 April 2020; Published: 15 April 2020 Abstract: Following the identification of the BCR-ABL1 (Breakpoint Cluster Region-ABelson murine Leukemia) fusion in chronic myelogenous leukemia, gene fusions generating chimeric oncoproteins have been recognized as common genomic structural variations in human malignancies. This is, in particular, a frequent mechanism in the oncogenic conversion of protein kinases. Gene fusion was the first mechanism identified for the oncogenic activation of the receptor tyrosine kinase RET (REarranged during Transfection), initially discovered in papillary thyroid carcinoma (PTC). More recently, the advent of highly sensitive massive parallel (next generation sequencing, NGS) sequencing of tumor DNA or cell-free (cfDNA) circulating tumor DNA, allowed for the detection of RET fusions in many other solid and hematopoietic malignancies. This review summarizes the role of RET fusions in the pathogenesis of human cancer. Keywords: kinase; tyrosine kinase inhibitor; targeted therapy; thyroid cancer 1. The RET Receptor RET (REarranged during Transfection) was initially isolated as a rearranged oncoprotein upon the transfection of a human lymphoma DNA [1]. -
Kinase Inhibitors: an Introduction
David Peters Baran Group Meeting Kinase Inhibitors: An Introduction 2/2/19 INTRODUCTION SIGNAL TRANSDUCTION KINASES ARE IMPORTANT IN HUMAN BIOLOGY/DISEASE: The process describing how a signal (chemical/physical) is transmitted through a - Kinases are a superfamily of proteins (5th largest in humans) cell ultimately resulting in a cellular response - 518 genes and 106 pseudogenes - Diverse in size, subunit structure, and cellular location RECEPTOR TRANSDUCERS EFFECTORS - ~260 residues make up their conserved catalytic core - dysregulation of kinases occurs in many diseases (cancer, inflamatory, degenerative, and autoimmune diseases) - signals can originate inside or outside the cell - 244 of the 518 map to disease loci - signals generally passed through a series of steps (signal transduction pathway) often consisting of multiple enzymes and messengers protein O - pathways open possibility for signal aplification (>1x106) kinase ATP + PROTEIN OH ADP + PROTEIN O P O - Extracellular signals transduced by RTKs, GPCR’s, guanlyl cyclases, or ligand-gated Ion channels O - Phosphorylation is the most prominent covalent modification/signal in KINASES INHIBITORS ARE IMPORTANT IN DISEASE THERAPY/RESEARCH: cellular regulation - currently 51 FDA approved small molecule kinase inhibitors - “converter enzymes” (protein kinases and phosphoprotein phosphorlyases) - 4 FDA approved antibody kinase inhibitors are regulated; conserve ATP/maintain desired target protein state - thousands of known inhibitors spanning the kinome - pathway ends by affecting biomolecule -

A Tyrosine Kinase Expression Signature Predicts the Post-Operative Clinical Outcome in Triple Negative Breast Cancers
cancers Article A Tyrosine Kinase Expression Signature Predicts the Post-Operative Clinical Outcome in Triple Negative Breast Cancers Alexandre de Nonneville 1, Pascal Finetti 2 , José Adelaide 2, Éric Lambaudie 3, Patrice Viens 1, Anthony Gonçalves 1 , Daniel Birnbaum 2, Emilie Mamessier 2 and François Bertucci 1,2,* 1 Department of Medical Oncology, Institut Paoli-Calmettes, Aix-Marseille Univ, CRCM, CNRS, INSERM, 13000 Marseille, France 2 Laboratory of Predictive Oncology, Centre de Recherche en Cancérologie de Marseille, Institut Paoli-Calmettes, Inserm UMR1068, CNRS UMR725, Aix-Marseille Université, 13000 Marseille, France 3 Department of Surgical Oncology, Institut Paoli-Calmettes, Aix-Marseille Univ, CNRS, INSERM, CRCM, 13000 Marseille, France * Correspondence: [email protected]; Tel.: +33-4-91-22-35-37; Fax: +33-4-91-22-36-70 Received: 15 July 2019; Accepted: 9 August 2019; Published: 13 August 2019 Abstract: Triple negative breast cancer (TNBC) represent 15% of breast cancers. Histoclinical features and marketed prognostic gene expression signatures (GES) failed to identify good- and poor-prognosis patients. Tyrosine kinases (TK) represent potential prognostic and/or therapeutic targets for TNBC. We sought to define a prognostic TK GES in a large series of TNBC. mRNA expression and histoclinical data of 6379 early BCs were collected from 16 datasets. We searched for a TK-based GES associated with disease-free survival (DFS) and tested its robustness in an independent validation set. A total of 1226 samples were TNBC. In the learning set of samples (N = 825), we identified a 13-TK GES associated with DFS. This GES was associated with cell proliferation and immune response. -

Functional Analysis of Somatic Mutations Affecting Receptor Tyrosine Kinase Family in Metastatic Colorectal Cancer
Author Manuscript Published OnlineFirst on March 29, 2019; DOI: 10.1158/1535-7163.MCT-18-0582 Author manuscripts have been peer reviewed and accepted for publication but have not yet been edited. Functional analysis of somatic mutations affecting receptor tyrosine kinase family in metastatic colorectal cancer Leslie Duplaquet1, Martin Figeac2, Frédéric Leprêtre2, Charline Frandemiche3,4, Céline Villenet2, Shéhérazade Sebda2, Nasrin Sarafan-Vasseur5, Mélanie Bénozène1, Audrey Vinchent1, Gautier Goormachtigh1, Laurence Wicquart6, Nathalie Rousseau3, Ludivine Beaussire5, Stéphanie Truant7, Pierre Michel8, Jean-Christophe Sabourin9, Françoise Galateau-Sallé10, Marie-Christine Copin1,6, Gérard Zalcman11, Yvan De Launoit1, Véronique Fafeur1 and David Tulasne1 1 Univ. Lille, CNRS, Institut Pasteur de Lille, UMR 8161 - M3T – Mechanisms of Tumorigenesis and Target Therapies, F-59000 Lille, France. 2 Univ. Lille, Plateau de génomique fonctionnelle et structurale, CHU Lille, F-59000 Lille, France 3 TCBN - Tumorothèque Caen Basse-Normandie, F-14000 Caen, France. 4 Réseau Régional de Cancérologie – OncoBasseNormandie – F14000 Caen – France. 5 Normandie Univ, UNIROUEN, Inserm U1245, IRON group, Rouen University Hospital, Normandy Centre for Genomic and Personalized Medicine, F-76000 Rouen, France. 6 Tumorothèque du C2RC de Lille, F-59037 Lille, France. 7 Department of Digestive Surgery and Transplantation, CHU Lille, Univ Lille, 2 Avenue Oscar Lambret, 59037, Lille Cedex, France. 8 Department of hepato-gastroenterology, Rouen University Hospital, Normandie Univ, UNIROUEN, Inserm U1245, IRON group, F-76000 Rouen, France. 9 Department of Pathology, Normandy University, INSERM 1245, Rouen University Hospital, F 76 000 Rouen, France. 10 Department of Pathology, MESOPATH-MESOBANK, Centre León Bérard, Lyon, France. 11 Thoracic Oncology Department, CIC1425/CLIP2 Paris-Nord, Hôpital Bichat-Claude Bernard, Paris, France. -

Clinical Activity of Ceritinib in ROS1-Rearranged Non-Small Cell Lung Cancer: Bench to Bedside Report LETTER Vivek Subbiaha,1, David S
LETTER Clinical activity of ceritinib in ROS1-rearranged non-small cell lung cancer: Bench to bedside report LETTER Vivek Subbiaha,1, David S. Honga, and Funda Meric-Bernstama We read with great interest the article by Davare et al. (1) profiling revealed a CD72-ROS1 rearrangement. The on structural insight of ROS1 tyrosine kinase inhibitors. patient was started on crizotinib at 250 mg orally twice Non-small cell lung cancer (NSCLC) is no longer a single daily and exhibited resolution of metastatic disease. disease but a collection of genetically heterogeneous The patient remained disease-free for 13 mo when tumors with different therapeutic options [e.g., aberra- CT-scan showed a relapse with two nodules in the tions in EGFR, BRAF, HER2/Neu, RET, anaplastic lym- right lower lobe. He underwent stereotactic radiation phoma kinase (ALK), and ROS1-rearranged tumors]. therapy. Imaging showed a treatment response but Each of these options has different clinical characteristics new pleural nodules. MRI brain scan showed new in- and therapeutic options with agents that have varying tracranial metastases. After undergoing gamma knife degrees of systemic activity. ROS1 gene rearrange- radiosurgery, the patient was enrolled in an ipilimumab ments define a distinct molecular subgroup of NSCLC. and radiation trial (NCT02239900). His disease pro- ROS1 rearrangement leads to constitutive ROS1 activa- gressed on ipilimumab. The patient was next enrolled tion and activity of crizotinib against ROS1-rearranged on the “Signature Trial,” a modular phase II study to link NSCLC was noted and crizotinib received regulatory targeted therapy to patients with pathway activated tu- approval for the treatment of ROS1 rearranged NSCLC mors; in this study patients whose tumors have aberra- (2). -

Lung Adenocarcinoma Therapeutic Implications
Patient Name Report Date Tumor Type Smith, James 10 January 2016 Lung adenocarcinoma Date of Birth 01 January 1950 Medical Facility Cancer Center Specimen Received 02 January 2016 Sex Male Ordering Physician Williams, Jane Specimen Site Liver FMI Case # TRF000000 Additional Recipient Not Given Date of Collection 03 January 2016 Medical Record # 100001 Medical Facility ID # 000001 Specimen Type Slide Specimen ID SID-00001 Pathologist Not Provided ABOUT THE TEST: FoundationOne™ is a next-generation sequencing (NGS) based assay that identifies genomic alterations within hundreds of cancer-related genes. PATIENT RESULTS TUMOR TYPE: LUNG ADENOCARCINOMA † 6 genomic alterations Genomic Alterations Identified ROS1 CD74-ROS1 fusion 3 therapies associated with potential clinical benefit CDK4 amplification MDM2 amplification 0 therapies associated with lack of response RICTOR amplification APC S688* 18 clinical trials FGF10 amplification Additional Disease-relevant Genes with No Reportable Alterations Identified† EGFR KRAS ALK BRAF MET RET ERBB2 † For a complete list of the genes assayed and performance specifications, please refer to the Appendix THERAPEUTIC IMPLICATIONS Genomic Alterations FDA-Approved Therapies FDA-Approved Therapies Potential Clinical Trials Detected (in patient’s tumor type) (in another tumor type) ROS1 Ceritinib None Yes, see clinical trials CD74-ROS1 fusion Crizotinib section CDK4 None Palbociclib Yes, see clinical trials amplification section MDM2 None None Yes, see clinical trials amplification section RICTOR None None Yes, see clinical trials amplification SAMPLEsection For more comprehensive information please log on to the Interactive Cancer Explorer™ To set up your Interactive Cancer Explorer account, contact your sales representative or call (888) 988-3639. Electronically Signed by Jeffrey S. -

CT Features and Disease Spread Patterns in ROS1-Rearranged Lung
www.nature.com/scientificreports OPEN CT features and disease spread patterns in ROS1‑rearranged lung adenocarcinomas: comparison with those of EGFR‑mutant or ALK‑rearranged lung adenocarcinomas Jung Han Woo1, Tae Jung Kim1*, Tae Sung Kim1 & Joungho Han2 The purpose of this study was to investigate the diferences in CT characteristics and disease spread patterns between ROS1‑rearranged adenocarcinomas and epidermal growth factor receptor (EGFR)‑mutant or anaplastic lymphoma kinase (ALK)‑rearranged adenocarcinomas. Patients with stage IIIb/IV adenocarcinoma with ROS1 rearrangement, EGFR mutations, or ALK rearrangement were retrospectively identifed. Two radiologists evaluated CT features and disease spread patterns. A multivariable logistic regression model was applied to determine the clinical and CT characteristics that can discriminate between ROS1‑rearranged and EGFR‑mutant or ALK‑ rearranged adenocarcinomas. A cohort of 169 patients was identifed (ROS1 = 23, EGFR = 120, and ALK = 26). Compared to EGFR‑mutant adenocarcinomas, ROS1‑rearranged adenocarcinomas were less likely to have air-bronchogram (p = 0.011) and pleural retraction (p = 0.048) and more likely to have pleural efusion (p = 0.025), pericardial metastases (p < 0.001), intrathoracic and extrathoracic nodal metastases (p = 0.047 and 0.023, respectively), and brain metastases (p = 0.017). Following multivariable analysis, age (OR = 1.06; 95% CI: 1.01, 1.12; p = 0.024), pericardial metastases (OR = 10.50; 95% CI: 2.10, 52.60; p = 0.005), and nodal metastases (OR = 8.55; 95% CI: 1.14, 62.52; p = 0.037) were found to be more common in ROS1‑rearranged tumors than in non‑ROS1‑rearranged tumors. ROS1‑rearranged adenocarcinomas appeared as solid tumors and were associated with young age, pericardial metastases and advanced nodal metastases relative to tumors with EGFR mutations or ALK rearrangement. -

Ephrin-A Binding and Epha Receptor Expression Delineate the Matrix Compartment of the Striatum
The Journal of Neuroscience, June 15, 1999, 19(12):4962–4971 Ephrin-A Binding and EphA Receptor Expression Delineate the Matrix Compartment of the Striatum L. Scott Janis, Robert M. Cassidy, and Lawrence F. Kromer Department of Cell Biology and Interdisciplinary Program in Neuroscience, Georgetown University Medical Center, Washington, DC 20007 The striatum integrates limbic and neocortical inputs to regulate matrix neurons. In situ hybridization for EphA RTKs reveals that sensorimotor and psychomotor behaviors. This function is de- the two different ligand binding patterns strictly match pendent on the segregation of striatal projection neurons into the mRNA expression patterns of EphA4 and EphA7. anatomical and functional components, such as the striosome Ligand–receptor binding assays indicate that ephrin-A1 and and matrix compartments. In the present study the association ephrin-A4 selectively bind EphA4 but not EphA7 in the lysates of ephrin-A cell surface ligands and EphA receptor tyrosine of striatal tissue. Conversely, ephrin-A2, ephrin-A3, and kinases (RTKs) with the organization of these compartments ephrin-A5 bind EphA7 but not EphA4. These observations im- was determined in postnatal rats. Ephrin-A1 and ephrin-A4 plicate selective interactions between ephrin-A molecules and selectively bind to EphA receptors on neurons restricted to the EphA RTKs as potential mechanisms for regulating the com- matrix compartment. Binding is absent from the striosomes, partmental organization of the striatum. which were identified by m-opioid