An in Silico Drug Repurposing for COVID-19[Version 1; Peer Review: 2
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
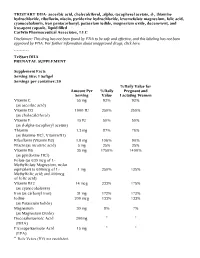
TRISTART DHA- Ascorbic Acid, Cholecalciferol, .Alpha
TRISTART DHA- ascorbic acid, cholecalciferol, .alpha.-tocopherol acetate, d-, thiamine hydrochloride, riboflavin, niacin, pyridoxine hydrochloride, levomefolate magnesium, folic acid, cyanocobalamin, iron pentacarbonyl, potassium iodide, magnesium oxide, doconexent, and icosapent capsule, liquid filled CarWin Pharmaceutical Associates, LLC Disclaimer: This drug has not been found by FDA to be safe and effective, and this labeling has not been approved by FDA. For further information about unapproved drugs, click here. ---------- TriStart DHA PRENATAL SUPPLEMENT Supplement Facts Serving Size: 1 Softgel Servings per container: 30 %Daily Value for Amount Per %Daily Pregnant and Serving Value Lactating Women Vitamin C 55 mg 92% 92% (as ascorbic acid) Vitamin D3 1000 IU 250% 250% (as cholecalciferol) Vitamin E 15 IU 50% 50% (as d-alpha-tocopheryl acetate) Thiamin 1.3 mg 87% 76% (as thiamine HCl, Vitamin B1) Riboflavin (Vitamin B2) 1.8 mg 106% 90% Niacin (as nicotinic acid) 5 mg 25% 25% Vitamin B6 35 mg 1750% 1400% (as pyridoxine HCl) Folate (as 630 mcg of L- Methylfolate Magnesium, molar equivalent to 600mcg of L- 1 mg 250% 125% Methylfolic acid; and 400mcg of folic acid) Vitamin B12 14 mcg 233% 175% (as cyanocobalamin) Iron (as carbonyl iron) 31 mg 172% 172% Iodine 200 mcg 133% 133% (as Potassium Iodide) Magnesium 30 mg 8% 7% (as Magnesium Oxide) Docosahexaenoic Acid 200mg * * (DHA) Eicosapentaenoate Acid 15 mg * * (EPA) * Daily Values (DV) not established. OTHER INGREDIENTS: Gelatin (bovine), Glycerin, Purified Water, Yellow Bees Wax, Caramel Powder, Soy Lecithin, Natural Orange Flavor, Ethyl Vanillin. Contains: Soy and Fish TriStart DHA™ Softgel capsules are dye free, lactose, gluten and sugar free. -

Enbrace® HR DESCRIPTION: INGREDIENTS
EnBrace® HR with DeltaFolate ™ [1 NF Units] [15 mg DFE Folate] ANTI-ANEMIA PREPARATION as extrinsic/intrinsic factor concentrate plus folate. Prescription Prenatal/Vitamin Drug For Therapeutic Use Multi-phasic Capsules (30ct bottle) NDC 64661-650-30 Rx Only [DRUG] GLUTEN-FREE DESCRIPTION: EnBrace® HR is an orally administered prescription prenatal/vitamin drug for therapeutic use formulated for female macrocytic anemia patients that are in need of treatment, and are under specific direction and monitoring of vitamin B12 and vitamin B9 status by a physician. EnBrace® HR is intended for women of childbearing age who are – or desire to become, pregnant regardless of lactation status. EnBrace® HR may be prescribed for women at risk of depression as a result of folate or cobalamin deficiency - including folate-induced postpartum depression, or are at risk of folate-induced birth defects such as may be found with spina bifida and other neural tube defects (NTDs). INGREDIENTS: Cobalamin intrinsic factor complex 1 NF Units* * National Formulary Units (“NF UNITS”) equivalent to 50 mcg of active coenzyme cobalamin (as cobamamide concentrate with intrinsic factor) ALSO CONTAINS: 1 Folinic acid (B9-vitamer) 2.5 mg + 1 Control-release, citrated folic acid, DHF (B9-Provitamin) 1 mg 2 Levomefolic acid (B9 & B12- cofactor) 5.23 mg 1 6 mg DFE folate (vitamin B9) 2 9 mg DFE l-methylfolate magnesium (molar equivalent). FUNCTIONAL EXCIPIENTS: 13.6 mg FeGC as ferrous glycine cysteinate (1.5 mg 3 3,4 elemental iron ) [colorant], 25 mg ascorbates (24 mg magnesium l-ascorbate, 1 mg zinc l-ascorbate) [antioxidant], at least 23.33 mg phospholipid-omega3 complex5 [marine lipids], 500 mcg betaine (trimethylglycine) [acidifier], 1 mg magnesium l-threonate [stabilizer]. -

Computational Antibiotics Book
Andrew V DeLong, Jared C Harris, Brittany S Larcart, Chandler B Massey, Chelsie D Northcutt, Somuayiro N Nwokike, Oscar A Otieno, Harsh M Patel, Mehulkumar P Patel, Pratik Pravin Patel, Eugene I Rowell, Brandon M Rush, Marc-Edwin G Saint-Louis, Amy M Vardeman, Felicia N Woods, Giso Abadi, Thomas J. Manning Computational Antibiotics Valdosta State University is located in South Georgia. Computational Antibiotics Index • Computational Details and Website Access (p. 8) • Acknowledgements (p. 9) • Dedications (p. 11) • Antibiotic Historical Introduction (p. 13) Introduction to Antibiotic groups • Penicillin’s (p. 21) • Carbapenems (p. 22) • Oxazolidines (p. 23) • Rifamycin (p. 24) • Lincosamides (p. 25) • Quinolones (p. 26) • Polypeptides antibiotics (p. 27) • Glycopeptide Antibiotics (p. 28) • Sulfonamides (p. 29) • Lipoglycopeptides (p. 30) • First Generation Cephalosporins (p. 31) • Cephalosporin Third Generation (p. 32) • Fourth-Generation Cephalosporins (p. 33) • Fifth Generation Cephalosporin’s (p. 34) • Tetracycline antibiotics (p. 35) Computational Antibiotics Antibiotics Covered (in alphabetical order) Amikacin (p. 36) Cefempidone (p. 98) Ceftizoxime (p. 159) Amoxicillin (p. 38) Cefepime (p. 100) Ceftobiprole (p. 161) Ampicillin (p. 40) Cefetamet (p. 102) Ceftoxide (p. 163) Arsphenamine (p. 42) Cefetrizole (p. 104) Ceftriaxone (p. 165) Azithromycin (p.44) Cefivitril (p. 106) Cefuracetime (p. 167) Aziocillin (p. 46) Cefixime (p. 108) Cefuroxime (p. 169) Aztreonam (p.48) Cefmatilen ( p. 110) Cefuzonam (p. 171) Bacampicillin (p. 50) Cefmetazole (p. 112) Cefalexin (p. 173) Bacitracin (p. 52) Cefodizime (p. 114) Chloramphenicol (p.175) Balofloxacin (p. 54) Cefonicid (p. 116) Cilastatin (p. 177) Carbenicillin (p. 56) Cefoperazone (p. 118) Ciprofloxacin (p. 179) Cefacetrile (p. 58) Cefoselis (p. 120) Clarithromycin (p. 181) Cefaclor (p. -

EMA/CVMP/158366/2019 Committee for Medicinal Products for Veterinary Use
Ref. Ares(2019)6843167 - 05/11/2019 31 October 2019 EMA/CVMP/158366/2019 Committee for Medicinal Products for Veterinary Use Advice on implementing measures under Article 37(4) of Regulation (EU) 2019/6 on veterinary medicinal products – Criteria for the designation of antimicrobials to be reserved for treatment of certain infections in humans Official address Domenico Scarlattilaan 6 ● 1083 HS Amsterdam ● The Netherlands Address for visits and deliveries Refer to www.ema.europa.eu/how-to-find-us Send us a question Go to www.ema.europa.eu/contact Telephone +31 (0)88 781 6000 An agency of the European Union © European Medicines Agency, 2019. Reproduction is authorised provided the source is acknowledged. Introduction On 6 February 2019, the European Commission sent a request to the European Medicines Agency (EMA) for a report on the criteria for the designation of antimicrobials to be reserved for the treatment of certain infections in humans in order to preserve the efficacy of those antimicrobials. The Agency was requested to provide a report by 31 October 2019 containing recommendations to the Commission as to which criteria should be used to determine those antimicrobials to be reserved for treatment of certain infections in humans (this is also referred to as ‘criteria for designating antimicrobials for human use’, ‘restricting antimicrobials to human use’, or ‘reserved for human use only’). The Committee for Medicinal Products for Veterinary Use (CVMP) formed an expert group to prepare the scientific report. The group was composed of seven experts selected from the European network of experts, on the basis of recommendations from the national competent authorities, one expert nominated from European Food Safety Authority (EFSA), one expert nominated by European Centre for Disease Prevention and Control (ECDC), one expert with expertise on human infectious diseases, and two Agency staff members with expertise on development of antimicrobial resistance . -

NOTES in Vitro Activities of Norfloxacin and Ciprofloxacin Against
ANTIMICROBIAL AGENTS AND CHEMOTHERAPY, July 1984, p. 94-96 Vol. 26, No. 1 0066-4804/84/070094-03$02.00/0 Copyright C 1984, American Society for Microbiology NOTES In Vitro Activities of Norfloxacin and Ciprofloxacin Against Mycobacterium tuberculosis, M. avium Complex, M. chelonei, M. fortuitum, and M. kansasii J. DOUGLAS GAY, DONALD R. DEYOUNG, AND GLENN D. ROBERTS* Section of Clinical Microbiology, Department of Laboratory Medicine, Mayo Clinic and Mayo Foundation, Rochester, Minnesota 55905 Received 28 November 1983/Accepted 4 April 1984 The activities of ciprofloxacin and norfloxacin against 100 mycobacteria isolates were studied in vitro by the 1% standard proportion method. Ciprofloxacin was more active against M. tuberculosis and M. fortuitum with MICs of 1.0 and 0.25 ,ug/ml, respectively, against 90% of isolates; norfloxacin had MICs of 8.0 and 2.0 ,ug/ml, respectively, against 90% of isolates. Nalidixic acid and other heterocyclic carbonic acid deriva- studied. The organisms were taken from the Mayo Clinic tives have been used primarily in the treatment of urinary stock culture collection, which included recent clinical iso- tract infections for many years. The compounds of this lates. Stock cultures were maintained on Middlebrook 7H10 general group include nalidixic acid, oxolinic acid, pipemidic agar slants (Difco Laboratories, Detroit, Mich.) and were acid, cinoxacin, and rosoxacin. Two new substances in this subcultured monthly. The identification of isolates was series which have been recently synthesized are norfloxacin based on standard biochemical tests (17) and gas-liquid (6) (1-ethyl-6-fluoro-1,4-dihydro-4-oxo-7-[ 1-piperazinyl ]-3- chromatography (16). -

Levomefolic Acid | Medchemexpress
Inhibitors Product Data Sheet Levomefolic acid • Agonists Cat. No.: HY-14781 CAS No.: 31690-09-2 Molecular Formula: C₂₀H₂₅N₇O₆ • Molecular Weight: 459.46 Screening Libraries Target: Endogenous Metabolite; Reactive Oxygen Species Pathway: Metabolic Enzyme/Protease; Immunology/Inflammation; NF-κB Storage: Powder -20°C 3 years 4°C 2 years In solvent -80°C 6 months -20°C 1 month SOLVENT & SOLUBILITY In Vitro H2O : 5 mg/mL (10.88 mM; Need ultrasonic) DMSO : < 1 mg/mL (insoluble or slightly soluble) Mass Solvent 1 mg 5 mg 10 mg Concentration Preparing 1 mM 2.1765 mL 10.8823 mL 21.7647 mL Stock Solutions 5 mM 0.4353 mL 2.1765 mL 4.3529 mL 10 mM 0.2176 mL 1.0882 mL 2.1765 mL Please refer to the solubility information to select the appropriate solvent. BIOLOGICAL ACTIVITY Description Levomefolic acid (5-MTHF) is the natural, active form of folic acid used at the cellular level for DNA reproduction, the cysteine cycle and the regulation of homocysteine among other functions. IC50 value: Target: Folate analogLevomefolic acid has been proposed for treatment of cardiovascular disease and advanced cancers such as breast and colorectal cancers. Levomefolic acid (5-MTHF) has the prominent antioxidant activity. A high dose of 5-MTHF or folic acid does not influence Natural killer (NK) cell function in vitro. CUSTOMER VALIDATION • J Mol Med (Berl). 2019 Aug;97(8):1183-1193. See more customer validations on www.MedChemExpress.com Page 1 of 2 www.MedChemExpress.com REFERENCES [1]. Hirsch S, et al. Natural killer cell cytotoxicity is not regulated by folic acid in vitro. -

WHO Report on Surveillance of Antibiotic Consumption: 2016-2018 Early Implementation ISBN 978-92-4-151488-0 © World Health Organization 2018 Some Rights Reserved
WHO Report on Surveillance of Antibiotic Consumption 2016-2018 Early implementation WHO Report on Surveillance of Antibiotic Consumption 2016 - 2018 Early implementation WHO report on surveillance of antibiotic consumption: 2016-2018 early implementation ISBN 978-92-4-151488-0 © World Health Organization 2018 Some rights reserved. This work is available under the Creative Commons Attribution- NonCommercial-ShareAlike 3.0 IGO licence (CC BY-NC-SA 3.0 IGO; https://creativecommons. org/licenses/by-nc-sa/3.0/igo). Under the terms of this licence, you may copy, redistribute and adapt the work for non- commercial purposes, provided the work is appropriately cited, as indicated below. In any use of this work, there should be no suggestion that WHO endorses any specific organization, products or services. The use of the WHO logo is not permitted. If you adapt the work, then you must license your work under the same or equivalent Creative Commons licence. If you create a translation of this work, you should add the following disclaimer along with the suggested citation: “This translation was not created by the World Health Organization (WHO). WHO is not responsible for the content or accuracy of this translation. The original English edition shall be the binding and authentic edition”. Any mediation relating to disputes arising under the licence shall be conducted in accordance with the mediation rules of the World Intellectual Property Organization. Suggested citation. WHO report on surveillance of antibiotic consumption: 2016-2018 early implementation. Geneva: World Health Organization; 2018. Licence: CC BY-NC-SA 3.0 IGO. Cataloguing-in-Publication (CIP) data. -

DENOVO- Levomefolic Acid Capsule Magna Pharmaceuticals, Inc
DENOVO- levomefolic acid capsule Magna Pharmaceuticals, Inc. ---------- Denovo Supplement Facts Serving Size: 1 capsule Servings per Container: 30 Each capsule contains the following: Folate [as 15,000mcg (as 15mg) DFE (dietary folate equivalents) L-5 MTHF (from L-5 methyltetrahydrofolate calcium salt)] Other Ingredients: Microcrystalline cellulose, capsule (gelatin,titanium dioxide, FD&C yellow #5, FD&C yellow #6), vegetable magnesium stearate, and silica Warning and Precautions Folic Acid, when administered in daily doses above 0.1mg may obscure the detection of B12 deficiency (specifically, the administration of folic acid may reverse the hematological manifestations of B12 deficiency, including pernicious anemia, while not addressing the neurological manifestations). 5- MTHF may be less likely than foli acid to mask vitamin B12 deficiency. Folate therapy alone is inadequate for the treatment of a B12 deficiency. A major depressive episode may be the inital presentation of bipolar disorder. It is generally believed, although not established in controlled trials, that treating such an episode with an antidepressant alone may increase the likelihood of a precipitation of mixed/manic episode in patients at risk for bipolar disorder. Denovo is not an antidepressant. However, 5-MTHF has been shown to enhance antidepressant effects of known antidepressants. Caution is recommended in patients with a history of bipolar illness. Patients with depressive symptoms should be adequately screened to determine if they are at risk for bipolar disorder since mood elevation in this population is possible. Denovo should always be used under medical supervision. Safe Handling Before using this product, tell your doctor or pharmacist of all of the products you use. -

(12) United States Patent (10) Patent No.: US 8,097,607 B2 Cabana Et Al
USO08097607B2 (12) United States Patent (10) Patent No.: US 8,097,607 B2 Cabana et al. (45) Date of Patent: *Jan. 17, 2012 (54) LOW DOSE RIFALAZIL COMPOSITIONS Emori et al., “Evaluation of in Vivo Therapeutic Efficacy of a New Benzoxazinorifamycin, KRM-1648, in SCID Mouse Model for Dis (76) Inventors: Bernard E. Cabana, Montgomery seminated Mycobacterium avium Complex Infection.” International Village, MD (US); Arthur F. Michaelis, Journal of Antimicrobial Agents 10(1):59 (1998). Devon, PA (US); Gary P. Magnant, Fujii et al., “In Vitro and In Vivo Antibacterial Activities of KRM Topsfield, MA (US); Chalom B. 1648 and KRM-1657, New Rifamycin Derivatives.” Antimicrobial Sayada, Luxembourg (LU) Agents and Chemotherapy 38: 1118, (1994). Gidoh et al., “Bactericidal Action at Low Doses of a New Rifamycin (*) Notice: Subject to any disclaimer, the term of this Derivative, 3'-hydroxy-5'-(4-isobutyl-1-piperazinyl) patent is extended or adjusted under 35 Benzoxazinorifamycin (KRM-1648) on Mycobacterium leprae U.S.C. 154(b) by 447 days. Inoculated into Ffootpads of Nude Mice.” Leprosy Review 63(4):319 This patent is Subject to a terminal dis (1992). claimer. Heep et al., “Detection of Rifabutin Resistance and Association of rpoB Mutation S with Resistance to Four Rifamycin Derivatives in Helicobacter pylori.” Journal of Clinical Microbiology & Infectious (21) Appl. No.: 10/668,792 Diseases 21:143 (2002). Hirara et al., “In Vitro and in Vivo Activities of the (22) Filed: Sep. 23, 2003 Benezoxazinorifamycin KRM-1648 Against Mycobacterium tuber (65) Prior Publication Data culosis,” Antimocrobial Agents and Chemotherapy 39 (10):2295 (1995). US 2004/O15784.0 A1 Aug. -

Pharmaceuticals As Environmental Contaminants
PharmaceuticalsPharmaceuticals asas EnvironmentalEnvironmental Contaminants:Contaminants: anan OverviewOverview ofof thethe ScienceScience Christian G. Daughton, Ph.D. Chief, Environmental Chemistry Branch Environmental Sciences Division National Exposure Research Laboratory Office of Research and Development Environmental Protection Agency Las Vegas, Nevada 89119 [email protected] Office of Research and Development National Exposure Research Laboratory, Environmental Sciences Division, Las Vegas, Nevada Why and how do drugs contaminate the environment? What might it all mean? How do we prevent it? Office of Research and Development National Exposure Research Laboratory, Environmental Sciences Division, Las Vegas, Nevada This talk presents only a cursory overview of some of the many science issues surrounding the topic of pharmaceuticals as environmental contaminants Office of Research and Development National Exposure Research Laboratory, Environmental Sciences Division, Las Vegas, Nevada A Clarification We sometimes loosely (but incorrectly) refer to drugs, medicines, medications, or pharmaceuticals as being the substances that contaminant the environment. The actual environmental contaminants, however, are the active pharmaceutical ingredients – APIs. These terms are all often used interchangeably Office of Research and Development National Exposure Research Laboratory, Environmental Sciences Division, Las Vegas, Nevada Office of Research and Development Available: http://www.epa.gov/nerlesd1/chemistry/pharma/image/drawing.pdfNational -

Surveillance of Antimicrobial Consumption in Europe 2013-2014 SURVEILLANCE REPORT
SURVEILLANCE REPORT SURVEILLANCE REPORT Surveillance of antimicrobial consumption in Europe in Europe consumption of antimicrobial Surveillance Surveillance of antimicrobial consumption in Europe 2013-2014 2012 www.ecdc.europa.eu ECDC SURVEILLANCE REPORT Surveillance of antimicrobial consumption in Europe 2013–2014 This report of the European Centre for Disease Prevention and Control (ECDC) was coordinated by Klaus Weist. Contributing authors Klaus Weist, Arno Muller, Ana Hoxha, Vera Vlahović-Palčevski, Christelle Elias, Dominique Monnet and Ole Heuer. Data analysis: Klaus Weist, Arno Muller and Ana Hoxha. Acknowledgements The authors would like to thank the ESAC-Net Disease Network Coordination Committee members (Marcel Bruch, Philippe Cavalié, Herman Goossens, Jenny Hellman, Susan Hopkins, Stephanie Natsch, Anna Olczak-Pienkowska, Ajay Oza, Arjana Tambić Andrasevic, Peter Zarb) and observers (Jane Robertson, Arno Muller, Mike Sharland, Theo Verheij) for providing valuable comments and scientific advice during the production of the report. All ESAC-Net participants and National Coordinators are acknowledged for providing data and valuable comments on this report. The authors also acknowledge Gaetan Guyodo, Catalin Albu and Anna Renau-Rosell for managing the data and providing technical support to the participating countries. Suggested citation: European Centre for Disease Prevention and Control. Surveillance of antimicrobial consumption in Europe, 2013‒2014. Stockholm: ECDC; 2018. Stockholm, May 2018 ISBN 978-92-9498-187-5 ISSN 2315-0955 -

European Surveillance of Healthcare-Associated Infections in Intensive Care Units
TECHNICAL DOCUMENT European surveillance of healthcare-associated infections in intensive care units HAI-Net ICU protocol Protocol version 1.02 www.ecdc.europa.eu ECDC TECHNICAL DOCUMENT European surveillance of healthcare- associated infections in intensive care units HAI-Net ICU protocol, version 1.02 This technical document of the European Centre for Disease Prevention and Control (ECDC) was coordinated by Carl Suetens. In accordance with the Staff Regulations for Officials and Conditions of Employment of Other Servants of the European Union and the ECDC Independence Policy, ECDC staff members shall not, in the performance of their duties, deal with a matter in which, directly or indirectly, they have any personal interest such as to impair their independence. This is version 1.02 of the HAI-Net ICU protocol. Differences between versions 1.01 (December 2010) and 1.02 are purely editorial. Suggested citation: European Centre for Disease Prevention and Control. European surveillance of healthcare- associated infections in intensive care units – HAI-Net ICU protocol, version 1.02. Stockholm: ECDC; 2015. Stockholm, March 2015 ISBN 978-92-9193-627-4 doi 10.2900/371526 Catalogue number TQ-04-15-186-EN-N © European Centre for Disease Prevention and Control, 2015 Reproduction is authorised, provided the source is acknowledged. TECHNICAL DOCUMENT HAI-Net ICU protocol, version 1.02 Table of contents Abbreviations ...............................................................................................................................................