Transcriptome Analysis Did Not Show Endogenous Stem Cell
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

1 Evidence for Gliadin Antibodies As Causative Agents in Schizophrenia
1 Evidence for gliadin antibodies as causative agents in schizophrenia. C.J.Carter PolygenicPathways, 20 Upper Maze Hill, Saint-Leonard’s on Sea, East Sussex, TN37 0LG [email protected] Tel: 0044 (0)1424 422201 I have no fax Abstract Antibodies to gliadin, a component of gluten, have frequently been reported in schizophrenia patients, and in some cases remission has been noted following the instigation of a gluten free diet. Gliadin is a highly immunogenic protein, and B cell epitopes along its entire immunogenic length are homologous to the products of numerous proteins relevant to schizophrenia (p = 0.012 to 3e-25). These include members of the DISC1 interactome, of glutamate, dopamine and neuregulin signalling networks, and of pathways involved in plasticity, dendritic growth or myelination. Antibodies to gliadin are likely to cross react with these key proteins, as has already been observed with synapsin 1 and calreticulin. Gliadin may thus be a causative agent in schizophrenia, under certain genetic and immunological conditions, producing its effects via antibody mediated knockdown of multiple proteins relevant to the disease process. Because of such homology, an autoimmune response may be sustained by the human antigens that resemble gliadin itself, a scenario supported by many reports of immune activation both in the brain and in lymphocytes in schizophrenia. Gluten free diets and removal of such antibodies may be of therapeutic benefit in certain cases of schizophrenia. 2 Introduction A number of studies from China, Norway, and the USA have reported the presence of gliadin antibodies in schizophrenia 1-5. Gliadin is a component of gluten, intolerance to which is implicated in coeliac disease 6. -
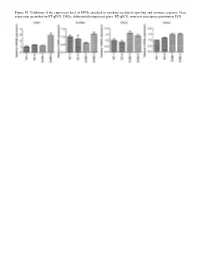
Figure S1. Validation of the Expression Level of Degs Enriched in Cytokine‑Mediated Signaling and Immune Response
Figure S1. Validation of the expression level of DEGs enriched in cytokine‑mediated signaling and immune response. Gene expression quantified by RT‑qPCR. DEGs, differentially expressed genes; RT‑qPCR, reverse‑transcription quantitative PCR. Figure S2. Validation of STAU1‑regulated AS events. IGV‑Sashimi plot revealed (A‑C) three A5SS AS events in three different genes. Reads distribution of each AS event was plotted in the left panel with the transcripts of each gene shown below. The sche‑ matic diagrams depict the structures of two AS events, AS1 (purple line) and AS2 (green line). The exon sequences are denoted by black boxes, the intron sequences by a horizontal line (right panel). RNA‑seq quantification and RT‑qPCR validation of ASEs are presented in the panels on the right. STAU1, double‑stranded RNA‑binding protein Staufen homolog 1; AS, alternative splicing; A5SS, alternative 5'splice site; RNA‑seq, RNA sequencing; RT‑qPCR, reverse‑transcription quantitative PCR. Error bars represent mean ± SEM. *P<0.05. Table SI. Primers used in gene validation experiments. IFIT2‑F CAGCCTACGGCAACTAAA IFIT2‑R GAGCCTTCTCAAAGCACA IFIT3‑F ACACCAAACAATGGCTAC IFIT3‑R TGGACAAACCCTCTAAAC OASL‑F AATGGTGACCGTGATGGG OASL‑R ACCTGAGGATGGAGCAGAG IFI27‑F TTCACTGCGGCGGGAATC IFI27‑R TGGCTGCTATGGAGGACGAG S1PR4‑F TGCTGAAGACGGTGCTGATG S1PR4‑R TGCGGAAGGAGTAGATGATGG CCL5‑F ACGACTGCTGGGTTGGAG CCL5‑R ACCCTGCTGCTTTGCCTA CCL2‑F CTAACCCAGAAACATCCAAT CCL2‑R GCTATGAGCAGCAGGCAC CD44‑F TGGAGGACAGAAAGCCAAGT CD44‑R TTCGCAATGAAACAATCAGTAG PLEKHG2‑M/As‑F CCAAAAGTAAGCCTGTCC PLEKHG2‑M‑R -

G Protein-Coupled Receptors
S.P.H. Alexander et al. The Concise Guide to PHARMACOLOGY 2015/16: G protein-coupled receptors. British Journal of Pharmacology (2015) 172, 5744–5869 THE CONCISE GUIDE TO PHARMACOLOGY 2015/16: G protein-coupled receptors Stephen PH Alexander1, Anthony P Davenport2, Eamonn Kelly3, Neil Marrion3, John A Peters4, Helen E Benson5, Elena Faccenda5, Adam J Pawson5, Joanna L Sharman5, Christopher Southan5, Jamie A Davies5 and CGTP Collaborators 1School of Biomedical Sciences, University of Nottingham Medical School, Nottingham, NG7 2UH, UK, 2Clinical Pharmacology Unit, University of Cambridge, Cambridge, CB2 0QQ, UK, 3School of Physiology and Pharmacology, University of Bristol, Bristol, BS8 1TD, UK, 4Neuroscience Division, Medical Education Institute, Ninewells Hospital and Medical School, University of Dundee, Dundee, DD1 9SY, UK, 5Centre for Integrative Physiology, University of Edinburgh, Edinburgh, EH8 9XD, UK Abstract The Concise Guide to PHARMACOLOGY 2015/16 provides concise overviews of the key properties of over 1750 human drug targets with their pharmacology, plus links to an open access knowledgebase of drug targets and their ligands (www.guidetopharmacology.org), which provides more detailed views of target and ligand properties. The full contents can be found at http://onlinelibrary.wiley.com/doi/ 10.1111/bph.13348/full. G protein-coupled receptors are one of the eight major pharmacological targets into which the Guide is divided, with the others being: ligand-gated ion channels, voltage-gated ion channels, other ion channels, nuclear hormone receptors, catalytic receptors, enzymes and transporters. These are presented with nomenclature guidance and summary information on the best available pharmacological tools, alongside key references and suggestions for further reading. -

Multi-Functionality of Proteins Involved in GPCR and G Protein Signaling: Making Sense of Structure–Function Continuum with In
Cellular and Molecular Life Sciences (2019) 76:4461–4492 https://doi.org/10.1007/s00018-019-03276-1 Cellular andMolecular Life Sciences REVIEW Multi‑functionality of proteins involved in GPCR and G protein signaling: making sense of structure–function continuum with intrinsic disorder‑based proteoforms Alexander V. Fonin1 · April L. Darling2 · Irina M. Kuznetsova1 · Konstantin K. Turoverov1,3 · Vladimir N. Uversky2,4 Received: 5 August 2019 / Revised: 5 August 2019 / Accepted: 12 August 2019 / Published online: 19 August 2019 © Springer Nature Switzerland AG 2019 Abstract GPCR–G protein signaling system recognizes a multitude of extracellular ligands and triggers a variety of intracellular signal- ing cascades in response. In humans, this system includes more than 800 various GPCRs and a large set of heterotrimeric G proteins. Complexity of this system goes far beyond a multitude of pair-wise ligand–GPCR and GPCR–G protein interactions. In fact, one GPCR can recognize more than one extracellular signal and interact with more than one G protein. Furthermore, one ligand can activate more than one GPCR, and multiple GPCRs can couple to the same G protein. This defnes an intricate multifunctionality of this important signaling system. Here, we show that the multifunctionality of GPCR–G protein system represents an illustrative example of the protein structure–function continuum, where structures of the involved proteins represent a complex mosaic of diferently folded regions (foldons, non-foldons, unfoldons, semi-foldons, and inducible foldons). The functionality of resulting highly dynamic conformational ensembles is fne-tuned by various post-translational modifcations and alternative splicing, and such ensembles can undergo dramatic changes at interaction with their specifc partners. -

Association of Dopaminergic/Gabaergic Genes with Attention Deficit Hyperactivity Disorder in Children
MOLECULAR MEDICINE REPORTS 6: 1093-1098, 2012 Association of dopaminergic/GABAergic genes with attention deficit hyperactivity disorder in children GUANG-XIN WANG1, YAN-HUI MA1, SHI-FU WANG1, GUANG-FANG REN2 and HUI GUO1 1Medical Institute of Paediatrics, Qilu Children's Hospital of Shandong University, Jinan, Shandong 250022; 2Department of Outpatients, Linshu People's Hospital of Shandong Province, Linshu, Shandong 276700, P.R. China Received March 16, 2012; Accepted August 1, 2012 DOI: 10.3892/mmr.2012.1028 Abstract. Attention deficit hyperactivity disorder (ADHD) is Although the etiology of ADHD is unknown, family, the most commonly diagnosed neurobehavioral disorder in twin and adoption studies have demonstrated that ADHD children and adolescents; however, its etiology is unknown. is highly heritable. It is widely accepted that several genes, In this study, we investigated the association of five poly- which include dopamine transporter gene (DAT1), dopamine morphisms in dopaminergic/GABAergic genes with ADHD D4 receptor gene (DRD4), synaptosomal-associated protein using polymerase chain reaction-restriction fragment length of 25 kDa gene (SNAP-25), DRD5, serotonin transporter polymorphism in a group of 54 children with ADHD and gene (5-HTT) and serotonin receptor 1B (5-HTR1B), each 67 healthy controls. The distribution of AA genotype and contributing a small fraction of the total genetic variance, A allele frequencies of rs5320 in the dopamine beta-hydroxy- are implicated in ADHD (3). Animal, pharmacological and lase gene in ADHD children differed significantly from that in imaging studies have indicated dopamine system dysfunc- healthy controls; however, no associations were found between tion in the pathogenesis of ADHD; therefore, dopaminergic four other polymorphisms in dopaminergic/GABAergic genes system genes may be candidate genes for ADHD (2). -

Understanding the Genetics and Neurobiological Pathways Behind Addiction (Review)
EXPERIMENTAL AND THERAPEUTIC MEDICINE 21: 544, 2021 Understanding the genetics and neurobiological pathways behind addiction (Review) ALEXANDRA POPESCU1, MARIA MARIAN1*, ANA MIRUNA DRĂGOI1* and RADU‑VIRGIL COSTEA2* 1Department of Psychiatry, ‘Prof. Dr. Alex. Obregia’ Clinical Hospital of Psychiatry, 041914 Bucharest; 2Department of General Surgery, ‘Carol Davila’ University of Medicine and Pharmacy, 020021 Bucharest, Romania Received January 15, 2021; Accepted February 15, 2021 DOI: 10.3892/etm.2021.9976 Abstract. The hypothesis issued by modern medicine states 1. Introduction that many diseases known to humans are genetically deter‑ mined, influenced or not by environmental factors, which is Substance‑related disorders are a set of behavioral, cognitive applicable to most psychiatric disorders as well. This article and physiological phenomena that occur after repeated use of a focuses on two pending questions regarding addiction: substance. These typically include: A strong desire to continue Why do some individuals become addicted while others do using a drug, difficulties in controlling its use, persistence in not? along with Is it a learned behavior or is it genetically using it although it has negative consequences, the use of the predefined? Recent data suggest that addiction is more than substance taking precedence over other activities and obliga‑ repeated exposure, it is the synchronicity between intrinsic tions, along with high tolerance and sometimes withdrawal (1). factors (genotype, sex, age, preexisting addictive disorder, New developments have altered the way we define addiction. or other mental illness), extrinsic factors (childhood, level of In this sense the Diagnostic and Statistical Manual of Mental education, socioeconomic status, social support, entourage, Disorders (5th ed.) (DSM‑5) (2) has changed the related chapter drug availability) and the nature of the addictive agent (phar‑ from ‘Substance‑Related Disorders’ to ‘Substance‑Related macokinetics, path of administration, psychoactive properties). -

Download (PDF)
Supplemental Information Biological and Pharmaceutical Bulletin Promoter Methylation Profiles between Human Lung Adenocarcinoma Multidrug Resistant A549/Cisplatin (A549/DDP) Cells and Its Progenitor A549 Cells Ruiling Guo, Guoming Wu, Haidong Li, Pin Qian, Juan Han, Feng Pan, Wenbi Li, Jin Li, and Fuyun Ji © 2013 The Pharmaceutical Society of Japan Table S1. Gene categories involved in biological functions with hypomethylated promoter identified by MeDIP-ChIP analysis in lung adenocarcinoma MDR A549/DDP cells compared with its progenitor A549 cells Different biological Genes functions transcription factor MYOD1, CDX2, HMX1, THRB, ARNT2, ZNF639, HOXD13, RORA, FOXO3, HOXD10, CITED1, GATA1, activity HOXC6, ZGPAT, HOXC8, ATOH1, FLI1, GATA5, HOXC4, HOXC5, PHTF1, RARB, MYST2, RARG, SIX3, FOXN1, ZHX3, HMG20A, SIX4, NR0B1, SIX6, TRERF1, DDIT3, ASCL1, MSX1, HIF1A, BAZ1B, MLLT10, FOXG1, DPRX, SHOX, ST18, CCRN4L, TFE3, ZNF131, SOX5, TFEB, MYEF2, VENTX, MYBL2, SOX8, ARNT, VDR, DBX2, FOXQ1, MEIS3, HOXA6, LHX2, NKX2-1, TFDP3, LHX6, EWSR1, KLF5, SMAD7, MAFB, SMAD5, NEUROG1, NR4A1, NEUROG3, GSC2, EN2, ESX1, SMAD1, KLF15, ZSCAN1, VAV1, GAS7, USF2, MSL3, SHOX2, DLX2, ZNF215, HOXB2, LASS3, HOXB5, ETS2, LASS2, DLX5, TCF12, BACH2, ZNF18, TBX21, E2F8, PRRX1, ZNF154, CTCF, PAX3, PRRX2, CBFA2T2, FEV, FOS, BARX1, PCGF2, SOX15, NFIL3, RBPJL, FOSL1, ALX1, EGR3, SOX14, FOXJ1, ZNF92, OTX1, ESR1, ZNF142, FOSB, MIXL1, PURA, ZFP37, ZBTB25, ZNF135, HOXC13, KCNH8, ZNF483, IRX4, ZNF367, NFIX, NFYB, ZBTB16, TCF7L1, HIC1, TSC22D1, TSC22D2, REXO4, POU3F2, MYOG, NFATC2, ENO1, -

A Bioinformatics Model of Human Diseases on the Basis Of
SUPPLEMENTARY MATERIALS A Bioinformatics Model of Human Diseases on the basis of Differentially Expressed Genes (of Domestic versus Wild Animals) That Are Orthologs of Human Genes Associated with Reproductive-Potential Changes Vasiliev1,2 G, Chadaeva2 I, Rasskazov2 D, Ponomarenko2 P, Sharypova2 E, Drachkova2 I, Bogomolov2 A, Savinkova2 L, Ponomarenko2,* M, Kolchanov2 N, Osadchuk2 A, Oshchepkov2 D, Osadchuk2 L 1 Novosibirsk State University, Novosibirsk 630090, Russia; 2 Institute of Cytology and Genetics, Siberian Branch of Russian Academy of Sciences, Novosibirsk 630090, Russia; * Correspondence: [email protected]. Tel.: +7 (383) 363-4963 ext. 1311 (M.P.) Supplementary data on effects of the human gene underexpression or overexpression under this study on the reproductive potential Table S1. Effects of underexpression or overexpression of the human genes under this study on the reproductive potential according to our estimates [1-5]. ↓ ↑ Human Deficit ( ) Excess ( ) # Gene NSNP Effect on reproductive potential [Reference] ♂♀ NSNP Effect on reproductive potential [Reference] ♂♀ 1 increased risks of preeclampsia as one of the most challenging 1 ACKR1 ← increased risk of atherosclerosis and other coronary artery disease [9] ← [3] problems of modern obstetrics [8] 1 within a model of human diseases using Adcyap1-knockout mice, 3 in a model of human health using transgenic mice overexpressing 2 ADCYAP1 ← → [4] decreased fertility [10] [4] Adcyap1 within only pancreatic β-cells, ameliorated diabetes [11] 2 within a model of human diseases -

Early During Myelomagenesis Alterations in DNA Methylation That
Myeloma Is Characterized by Stage-Specific Alterations in DNA Methylation That Occur Early during Myelomagenesis This information is current as Christoph J. Heuck, Jayesh Mehta, Tushar Bhagat, Krishna of September 23, 2021. Gundabolu, Yiting Yu, Shahper Khan, Grigoris Chrysofakis, Carolina Schinke, Joseph Tariman, Eric Vickrey, Natalie Pulliam, Sangeeta Nischal, Li Zhou, Sanchari Bhattacharyya, Richard Meagher, Caroline Hu, Shahina Maqbool, Masako Suzuki, Samir Parekh, Frederic Reu, Ulrich Steidl, John Greally, Amit Verma and Seema B. Downloaded from Singhal J Immunol 2013; 190:2966-2975; Prepublished online 13 February 2013; doi: 10.4049/jimmunol.1202493 http://www.jimmunol.org/content/190/6/2966 http://www.jimmunol.org/ Supplementary http://www.jimmunol.org/content/suppl/2013/02/13/jimmunol.120249 Material 3.DC1 References This article cites 38 articles, 15 of which you can access for free at: http://www.jimmunol.org/content/190/6/2966.full#ref-list-1 by guest on September 23, 2021 Why The JI? Submit online. • Rapid Reviews! 30 days* from submission to initial decision • No Triage! Every submission reviewed by practicing scientists • Fast Publication! 4 weeks from acceptance to publication *average Subscription Information about subscribing to The Journal of Immunology is online at: http://jimmunol.org/subscription Permissions Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Email Alerts Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts The Journal of Immunology is published twice each month by The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2013 by The American Association of Immunologists, Inc. -

GPCR Expression Profiles Were Determined Using
Supplemental Figures and Tables for Tischner et al., 2017 Supplemental Figure 1: GPCR expression profiles were determined using the NanoString nCounter System in 250 ng of pooled cell RNA obtained from freshly isolated CD4 T cells from naïve lymph nodes (CD4ln), spinal cord infiltrating CD4 T cells at peak EAE disease (CD4sc), and primary lung endothelial cells (luEC). Supplemental Figure 2: Array design and quality controls. A, Sorted leukocytes or endothelial cells were subjected to single‐cell expression analysis and re‐evaluated based on the expression of various identity‐defining genes. B, Expression of identity‐defining and quality control genes after deletion of contaminating or reference gene‐negative cells. Expression data are calculated as 2(Limit of detection(LoD) Ct – sample Ct) ; LoD Ct was set to 24. Supplemental Figure 3: Overview over GPCR expression frequencies in different freshly isolated immune cell populations and spinal cord endothelial cells as determined by single cell RT‐PCR. Abbreviations: CD4ln‐Tcon/CD4ln‐Treg, conventional (con) and regulatory (reg) CD4 T cells from lymph nodes (CD4ln) of naïve mice; CD4dr/CD4sc, CD4 T cells from draining lymph nodes (dr) or spinal cord (sc) at peak EAE disease; CD4spn2D/ CD4spn2DTh1/ CD4spn2DTh17, splenic CD4 T cells from 2D2 T cell receptor transgenic mice before (2D) and after in vitro differentiation towards Th1 (2DTh1) or Th17 (2DTh17); MonoSpn, splenic monocytes; CD11b_sc, spinal cord infiltrating CD11b‐ positive cells; sc_microglia, Ccr2neg,Cx3cr1pos microglia from spinal cord at peak disease; sc_macrophages, CCr2pos;Cx3cr1lo/neg macrophages from spinal cord at peak disease; BMDM_M1/BMDM_M2, bone marrow‐derived macrophages differentiated towards M1 or M2; ECscN and ECscEAE, spinal cord endothelial cells from naïve mice (N) and at peak EAE disease (EAE); SMC, smooth muscle cells from various vessel types (included as positive control to ascertain primer functionality). -

USP8 Mutations in Corticotroph Adenomas Determine a Distinct Gene Expression Profile Irrespective of Functional Tumour Status
6 181 M Bujko, P Kober and Transcriptomic profiling in 181:6 615–627 Clinical Study others corticotrophinomas USP8 mutations in corticotroph adenomas determine a distinct gene expression profile irrespective of functional tumour status Mateusz Bujko1,*, Paulina Kober1,*, Joanna Boresowicz1, Natalia Rusetska1, Agnieszka Paziewska2,6, Michalina Dąbrowska2, Agata Piaścik3, Monika Pękul3, Grzegorz Zieliński4, Jacek Kunicki5, Wiesław Bonicki5, Jerzy Ostrowski2,6, Janusz A Siedlecki1 and Maria Maksymowicz3 1Department of Molecular and Translational Oncology, 2Department of Genetics, 3Department of Pathology and Laboratory Diagnostics, Maria Sklodowska-Curie Institute – Oncology Center, Warsaw, Poland, 4Department of Neurosurgery, Military Institute of Medicine, Warsaw, Poland, 5Department of Neurosurgery, Maria Sklodowska-Curie Institute – Correspondence Oncology Center, Warsaw, Poland, and 6Department of Gastroenterology, Hepatology and Clinical Oncology, Medical should be addressed Center for Postgraduate Education, Warsaw, Poland to M Maksymowicz *(M Bujko and P Kober contributed equally to this work) Email [email protected] Abstract Objective: Pituitary corticotroph adenomas commonly cause Cushing’s disease (CD) but part of these tumours are hormonally inactive (silent corticotroph adenomas, SCA). USP8 mutations are well-known driver mutations in corticotrophinomas. Differences in transcriptomic profiles between functioning and silent tumours or tumours with different USP8 status have not been investigated. Design and methods: Forty-eight patients (28 CD, 20 SCA) were screened for USP8 mutations with Sanger sequencing. Twenty-four patients were included in transcriptomic profiling with Ampliseq Transcriptome Human Gene Expression Core Panel. The entire patients group was included in qRT-PCR analysis of selected genes expression. Immunohistochemistry was used for visualization of selected protein. European Journal of Endocrinology Results: We found USP8 mutation in 15 patients with CD and 4 SCAs. -

Altered Transcription of Glutamatergic and Glycinergic Receptors in Spinal
European Journal of Neuroscience Altered transcription of glutamatergic and glycinergic receptors in spinal cord dorsal horn following spinal cord transection is minimally affected by passive exercise of the hindlimbs For Peer Review Journal: European Journal of Neuroscience Manuscript ID Draft Manuscript Type: Research Report Date Submitted by the Author: n/a Complete List of Authors: Chopek, Jeremy; Dalhousie University, Medical Neuroscience; University of Manitoba College of Medicine, Physiology and Pathophysiology MacDonell, Chris; University of Manitoba College of Medicine, Physiology and Pathophysiology Shepard, Patricia; University of Manitoba College of Medicine, Physiology and Pathophysiology Gardiner, Kalan; University of Manitoba College of Medicine, Physiology and Pathophysiology Gardiner, PF; University of Manitoba, Physiology and Pathophysiology Key Words: Spinal cord injury, gene expression, dorsal horn, exercise Page 5 of 32 European Journal of Neuroscience Dorsal horn gene expression following STx Journal Section: Molecular and synaptic mechanisms Title: Altered transcription of glutamatergic and glycinergic receptors in spinal cord dorsal horn following spinal cord transection is minimally affected by passive exercise of the hindlimbs Authors: Jeremy W.For Chopek, PeerChristopher W. Review MacDonell, Patricia C. Shepard, Kalan R. Gardiner, Phillip F Gardiner Affiliations: Spinal Cord Research Centre, Department of Physiology and Pathophysiology, Rady Faculty of Health, University of Manitoba, Winnipeg, MB, R3E 0J9 Running