In Vitro Profiling of Orphan G Protein Coupled Receptor (GPCR)
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Strategies to Increase ß-Cell Mass Expansion
This electronic thesis or dissertation has been downloaded from the King’s Research Portal at https://kclpure.kcl.ac.uk/portal/ Strategies to increase -cell mass expansion Drynda, Robert Lech Awarding institution: King's College London The copyright of this thesis rests with the author and no quotation from it or information derived from it may be published without proper acknowledgement. END USER LICENCE AGREEMENT Unless another licence is stated on the immediately following page this work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International licence. https://creativecommons.org/licenses/by-nc-nd/4.0/ You are free to copy, distribute and transmit the work Under the following conditions: Attribution: You must attribute the work in the manner specified by the author (but not in any way that suggests that they endorse you or your use of the work). Non Commercial: You may not use this work for commercial purposes. No Derivative Works - You may not alter, transform, or build upon this work. Any of these conditions can be waived if you receive permission from the author. Your fair dealings and other rights are in no way affected by the above. Take down policy If you believe that this document breaches copyright please contact [email protected] providing details, and we will remove access to the work immediately and investigate your claim. Download date: 02. Oct. 2021 Strategies to increase β-cell mass expansion A thesis submitted by Robert Drynda For the degree of Doctor of Philosophy from King’s College London Diabetes Research Group Division of Diabetes & Nutritional Sciences Faculty of Life Sciences & Medicine King’s College London 2017 Table of contents Table of contents ................................................................................................. -

Chimeric Gpcrs Mimic Distinct Signaling Pathways and Modulate Microglia Responses
bioRxiv preprint doi: https://doi.org/10.1101/2021.06.21.449162; this version posted June 21, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-ND 4.0 International license. Chimeric GPCRs mimic distinct signaling pathways and modulate microglia responses Rouven Schulz, Medina Korkut-Demirbaş, Gloria Colombo, Sandra Siegert† Author affiliation: Institute of Science and Technology (IST) Austria, Am Campus 1, 3400 Klosterneuburg, Austria † Corresponding author: [email protected] Keywords: G protein coupled receptor (GPCR), DREADD, β2-adrenergic receptor (β2AR/ADRB2), microglia, inflammation, GPR65, GPR109A/HCAR2 Abstract G protein-coupled receptors (GPCRs) regulate multiple processes ranging from cell growth and immune responses to neuronal signal transmission. However, ligands for many GPCRs remain unknown, suffer from off-target effects or have poor bioavailability. Additional challenges exist to dissect cell type-specific responses when the same GPCR is expressed on different cells within the body. Here, we overcome these limitations by engineering DREADD-based GPCR chimeras that selectively bind their agonist clozapine-N-oxide (CNO) and mimic a GPCR-of-interest. We show that the chimeric DREADD-β2-adrenergic receptor (β2AR/ADRB2) triggers comparable responses to levalbuterol on second messenger and kinase activity, post-translational modifications, and protein-protein interactions. Moreover, we successfully recapitulate β2AR-mediated filopodia formation in microglia, a β2AR-expressing immune cell that can drive inflammation in the nervous system. To further dissect microglial inflammation, we compared DREADD-β2AR with two additionally designed DREADD-based chimeras mimicking GPR65 and GPR109A/HCAR2, both enriched in microglia. -

1 Evidence for Gliadin Antibodies As Causative Agents in Schizophrenia
1 Evidence for gliadin antibodies as causative agents in schizophrenia. C.J.Carter PolygenicPathways, 20 Upper Maze Hill, Saint-Leonard’s on Sea, East Sussex, TN37 0LG [email protected] Tel: 0044 (0)1424 422201 I have no fax Abstract Antibodies to gliadin, a component of gluten, have frequently been reported in schizophrenia patients, and in some cases remission has been noted following the instigation of a gluten free diet. Gliadin is a highly immunogenic protein, and B cell epitopes along its entire immunogenic length are homologous to the products of numerous proteins relevant to schizophrenia (p = 0.012 to 3e-25). These include members of the DISC1 interactome, of glutamate, dopamine and neuregulin signalling networks, and of pathways involved in plasticity, dendritic growth or myelination. Antibodies to gliadin are likely to cross react with these key proteins, as has already been observed with synapsin 1 and calreticulin. Gliadin may thus be a causative agent in schizophrenia, under certain genetic and immunological conditions, producing its effects via antibody mediated knockdown of multiple proteins relevant to the disease process. Because of such homology, an autoimmune response may be sustained by the human antigens that resemble gliadin itself, a scenario supported by many reports of immune activation both in the brain and in lymphocytes in schizophrenia. Gluten free diets and removal of such antibodies may be of therapeutic benefit in certain cases of schizophrenia. 2 Introduction A number of studies from China, Norway, and the USA have reported the presence of gliadin antibodies in schizophrenia 1-5. Gliadin is a component of gluten, intolerance to which is implicated in coeliac disease 6. -

Viewed Under 23 (B) Or 203 (C) fi M M Male Cko Mice, and Largely Unaffected Magni Cation; Scale Bars, 500 M (B) and 50 M (C)
BRIEF COMMUNICATION www.jasn.org Renal Fanconi Syndrome and Hypophosphatemic Rickets in the Absence of Xenotropic and Polytropic Retroviral Receptor in the Nephron Camille Ansermet,* Matthias B. Moor,* Gabriel Centeno,* Muriel Auberson,* † † ‡ Dorothy Zhang Hu, Roland Baron, Svetlana Nikolaeva,* Barbara Haenzi,* | Natalya Katanaeva,* Ivan Gautschi,* Vladimir Katanaev,*§ Samuel Rotman, Robert Koesters,¶ †† Laurent Schild,* Sylvain Pradervand,** Olivier Bonny,* and Dmitri Firsov* BRIEF COMMUNICATION *Department of Pharmacology and Toxicology and **Genomic Technologies Facility, University of Lausanne, Lausanne, Switzerland; †Department of Oral Medicine, Infection, and Immunity, Harvard School of Dental Medicine, Boston, Massachusetts; ‡Institute of Evolutionary Physiology and Biochemistry, St. Petersburg, Russia; §School of Biomedicine, Far Eastern Federal University, Vladivostok, Russia; |Services of Pathology and ††Nephrology, Department of Medicine, University Hospital of Lausanne, Lausanne, Switzerland; and ¶Université Pierre et Marie Curie, Paris, France ABSTRACT Tight control of extracellular and intracellular inorganic phosphate (Pi) levels is crit- leaves.4 Most recently, Legati et al. have ical to most biochemical and physiologic processes. Urinary Pi is freely filtered at the shown an association between genetic kidney glomerulus and is reabsorbed in the renal tubule by the action of the apical polymorphisms in Xpr1 and primary fa- sodium-dependent phosphate transporters, NaPi-IIa/NaPi-IIc/Pit2. However, the milial brain calcification disorder.5 How- molecular identity of the protein(s) participating in the basolateral Pi efflux remains ever, the role of XPR1 in the maintenance unknown. Evidence has suggested that xenotropic and polytropic retroviral recep- of Pi homeostasis remains unknown. Here, tor 1 (XPR1) might be involved in this process. Here, we show that conditional in- we addressed this issue in mice deficient for activation of Xpr1 in the renal tubule in mice resulted in impaired renal Pi Xpr1 in the nephron. -

Edinburgh Research Explorer
Edinburgh Research Explorer International Union of Basic and Clinical Pharmacology. LXXXVIII. G protein-coupled receptor list Citation for published version: Davenport, AP, Alexander, SPH, Sharman, JL, Pawson, AJ, Benson, HE, Monaghan, AE, Liew, WC, Mpamhanga, CP, Bonner, TI, Neubig, RR, Pin, JP, Spedding, M & Harmar, AJ 2013, 'International Union of Basic and Clinical Pharmacology. LXXXVIII. G protein-coupled receptor list: recommendations for new pairings with cognate ligands', Pharmacological reviews, vol. 65, no. 3, pp. 967-86. https://doi.org/10.1124/pr.112.007179 Digital Object Identifier (DOI): 10.1124/pr.112.007179 Link: Link to publication record in Edinburgh Research Explorer Document Version: Publisher's PDF, also known as Version of record Published In: Pharmacological reviews Publisher Rights Statement: U.S. Government work not protected by U.S. copyright General rights Copyright for the publications made accessible via the Edinburgh Research Explorer is retained by the author(s) and / or other copyright owners and it is a condition of accessing these publications that users recognise and abide by the legal requirements associated with these rights. Take down policy The University of Edinburgh has made every reasonable effort to ensure that Edinburgh Research Explorer content complies with UK legislation. If you believe that the public display of this file breaches copyright please contact [email protected] providing details, and we will remove access to the work immediately and investigate your claim. Download date: 02. Oct. 2021 1521-0081/65/3/967–986$25.00 http://dx.doi.org/10.1124/pr.112.007179 PHARMACOLOGICAL REVIEWS Pharmacol Rev 65:967–986, July 2013 U.S. -

Supplementary Data
Supplemental Data A novel mouse model of X-linked nephrogenic diabetes insipidus: Phenotypic analysis and therapeutic implications Jian Hua Li, Chung-Lin Chou, Bo Li, Oksana Gavrilova, Christoph Eisner, Jürgen Schnermann, Stasia A. Anderson, Chu-Xia Deng, Mark A. Knepper, and Jürgen Wess Supplemental Methods Metabolic cage studies. Animals were maintained in mouse metabolic cages (Hatteras Instruments, Cary, NC) under controlled temperature and light conditions (12 hr light and dark cycles). Mice received a fixed daily ration of 6.5 g of gelled diet per 20 g of body weight per day. The gelled diet was composed of 4 g of Basal Diet 5755 (Test Diet, Richmond, IN), 2.5 ml of deionized water, and 65 mg agar. Preweighted drinking water was provided ad libitum during the course of the study. Mice were acclimated in the metabolic cages for 1-2 days. Urine was collected under mineral oil in preweighted collection vials for successive 24 hr periods. Analysis of GPCR expression in mouse IMCD cells via TaqMan real-time qRT-PCR. Total RNA prepared from mouse IMCD tubule suspensions was reverse transcribed as described under Experimental Procedures. Tissues from ten 10-week old C57BL/6 WT mice were collected and pooled for each individual experiment. cDNA derived from 640 ng of RNA was mixed with an equal volume of TaqMan gene expression 2 x master mix (Applied Biosystems, Foster City, CA). 100 μl-aliquots of this mixture (corresponding to 80 ng of RNA) were added to each of the 8 fill ports of a 384-well plate of a mouse GPCR array panel (Applied Biosystems). -

Supporting Online Material
1 2 3 4 5 6 7 Supplementary Information for 8 9 Fractalkine-induced microglial vasoregulation occurs within the retina and is altered early in diabetic 10 retinopathy 11 12 *Samuel A. Mills, *Andrew I. Jobling, *Michael A. Dixon, Bang V. Bui, Kirstan A. Vessey, Joanna A. Phipps, 13 Ursula Greferath, Gene Venables, Vickie H.Y. Wong, Connie H.Y. Wong, Zheng He, Flora Hui, James C. 14 Young, Josh Tonc, Elena Ivanova, Botir T. Sagdullaev, Erica L. Fletcher 15 * Joint first authors 16 17 Corresponding author: 18 Prof. Erica L. Fletcher. Department of Anatomy & Neuroscience. The University of Melbourne, Grattan St, 19 Parkville 3010, Victoria, Australia. 20 Email: [email protected] ; Tel: +61-3-8344-3218; Fax: +61-3-9347-5219 21 22 This PDF file includes: 23 24 Supplementary text 25 Figures S1 to S10 26 Tables S1 to S7 27 Legends for Movies S1 to S2 28 SI References 29 30 Other supplementary materials for this manuscript include the following: 31 32 Movies S1 to S2 33 34 35 36 1 1 Supplementary Information Text 2 Materials and Methods 3 Microglial process movement on retinal vessels 4 Dark agouti rats were anaesthetized, injected intraperitoneally with rhodamine B (Sigma-Aldrich) to label blood 5 vessels and retinal explants established as described in the main text. Retinal microglia were labelled with Iba-1 6 and imaging performed on an inverted confocal microscope (Leica SP5). Baseline images were taken for 10 7 minutes, followed by the addition of PBS (10 minutes) and then either fractalkine or fractalkine + candesartan 8 (10 minutes) using concentrations outlined in the main text. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

The Habenular G-Protein–Coupled Receptor 151 Regulates Synaptic Plasticity and Nicotine Intake
The habenular G-protein–coupled receptor 151 regulates synaptic plasticity and nicotine intake Beatriz Antolin-Fontesa, Kun Lia, Jessica L. Ablesa,b,c, Michael H. Riada, Andreas Görlicha, Maya Williamsb, Cuidong Wanga, Sylvia M. Lipforda, Maria Daob, Jianxi Liud, Henrik Molinae, Nathaniel Heintza,1, Paul J. Kennyb,d, and Ines Ibañez-Tallona1 aLaboratory of Molecular Biology, The Rockefeller University, New York, NY 10065; bNash Family Department of Neuroscience, Icahn School of Medicine at Mount Sinai, New York, NY 10029-6574; cDepartment of Psychiatry, Icahn School of Medicine at Mount Sinai, New York, NY 10029-6574; dDepartment of Pharmacology and Systems Therapeutics, Icahn School of Medicine at Mount Sinai, New York, NY 10029-6574; and eProteomics Resource Center, The Rockefeller University, New York, NY 10065 Contributed by Nathaniel Heintz, January 17, 2020 (sent for review September 22, 2019; reviewed by Ana Belén Elgoyhen and Gord Fishell) The habenula, an ancient small brain area in the epithalamus, domains corelease acetylcholine and glutamate, which activate densely expresses nicotinic acetylcholine receptors and is critical postsynaptic receptors via volume and wired transmission, re- for nicotine intake and aversion. As such, identification of spectively (10, 11). Some of the highest densities of nicotinic strategies to manipulate habenular activity may yield approaches acetylcholine receptors (nAChRs) in the brain are detected in the to treat nicotine addiction. Here we show that GPR151, an orphan MHb–IPN axis (12–15), especially of α5, α3, and β4nAChR G-protein–coupled receptor (GPCR) highly enriched in the habenula subunits. Little was known about the MHb in regulating the mo- of humans and rodents, is expressed at presynaptic membranes and tivational properties of nicotine until human genetics studies synaptic vesicles and associates with synaptic components control- established a strong association between genetic variants in the ling vesicle release and ion transport. -

G Protein-Coupled Receptors in Stem Cell Maintenance and Somatic Reprogramming to Pluripotent Or Cancer Stem Cells
BMB Reports - Manuscript Submission Manuscript Draft Manuscript Number: BMB-14-250 Title: G protein-coupled receptors in stem cell maintenance and somatic reprogramming to pluripotent or cancer stem cells Article Type: Mini Review Keywords: G protein-coupled receptors; stem cell maintenance; somatic reprogramming; cancer stem cells; pluripotent stem cell Corresponding Author: Ssang-Goo Cho Authors: Ssang-Goo Cho1,*, Hye Yeon Choi1, Subbroto Kumar Saha1, Kyeongseok Kim1, Sangsu Kim1, Gwang-Mo Yang1, BongWoo Kim1, Jin-hoi Kim1 Institution: 1Department of Animal Biotechnology, Animal Resources Research Center, and Incurable Disease Animal Model and Stem Cell Institute (IDASI), Konkuk University, 120 Neungdong-ro, Gwangjin-gu, Seoul 143-701, Republic of Korea, UNCORRECTED PROOF 1 G protein-coupled receptors in stem cell maintenance and somatic reprogramming to 2 pluripotent or cancer stem cells 3 4 Hye Yeon Choi, Subbroto Kumar Saha, Kyeongseok Kim, Sangsu Kim, Gwang-Mo 5 Yang, BongWoo Kim, Jin-hoi Kim, and Ssang-Goo Cho 6 7 Department of Animal Biotechnology, Animal Resources Research Center, and 8 Incurable Disease Animal Model and Stem Cell Institute (IDASI), Konkuk University, 9 120 Neungdong-ro, Gwangjin-gu, Seoul 143-701, Republic of Korea 10 * 11 Address correspondence to Ssang-Goo Cho, Department of Animal Biotechnology and 12 Animal Resources Research Center. Konkuk University, 120 Neungdong-ro, Gwangjin- 13 gu, Seoul 143-701, Republic of Korea. Tel: 82-2-450-4207, Fax: 82-2-450-1044, E-mail: 14 [email protected] 15 16 17 18 19 1 UNCORRECTED PROOF 20 Abstract 21 The G protein-coupled receptors (GPCRs) compose the third largest gene family in the 22 human genome, representing more than 800 distinct genes and 3–5% of the human genome. -
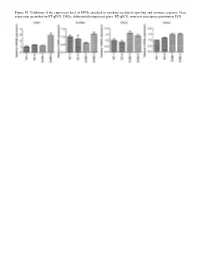
Figure S1. Validation of the Expression Level of Degs Enriched in Cytokine‑Mediated Signaling and Immune Response
Figure S1. Validation of the expression level of DEGs enriched in cytokine‑mediated signaling and immune response. Gene expression quantified by RT‑qPCR. DEGs, differentially expressed genes; RT‑qPCR, reverse‑transcription quantitative PCR. Figure S2. Validation of STAU1‑regulated AS events. IGV‑Sashimi plot revealed (A‑C) three A5SS AS events in three different genes. Reads distribution of each AS event was plotted in the left panel with the transcripts of each gene shown below. The sche‑ matic diagrams depict the structures of two AS events, AS1 (purple line) and AS2 (green line). The exon sequences are denoted by black boxes, the intron sequences by a horizontal line (right panel). RNA‑seq quantification and RT‑qPCR validation of ASEs are presented in the panels on the right. STAU1, double‑stranded RNA‑binding protein Staufen homolog 1; AS, alternative splicing; A5SS, alternative 5'splice site; RNA‑seq, RNA sequencing; RT‑qPCR, reverse‑transcription quantitative PCR. Error bars represent mean ± SEM. *P<0.05. Table SI. Primers used in gene validation experiments. IFIT2‑F CAGCCTACGGCAACTAAA IFIT2‑R GAGCCTTCTCAAAGCACA IFIT3‑F ACACCAAACAATGGCTAC IFIT3‑R TGGACAAACCCTCTAAAC OASL‑F AATGGTGACCGTGATGGG OASL‑R ACCTGAGGATGGAGCAGAG IFI27‑F TTCACTGCGGCGGGAATC IFI27‑R TGGCTGCTATGGAGGACGAG S1PR4‑F TGCTGAAGACGGTGCTGATG S1PR4‑R TGCGGAAGGAGTAGATGATGG CCL5‑F ACGACTGCTGGGTTGGAG CCL5‑R ACCCTGCTGCTTTGCCTA CCL2‑F CTAACCCAGAAACATCCAAT CCL2‑R GCTATGAGCAGCAGGCAC CD44‑F TGGAGGACAGAAAGCCAAGT CD44‑R TTCGCAATGAAACAATCAGTAG PLEKHG2‑M/As‑F CCAAAAGTAAGCCTGTCC PLEKHG2‑M‑R -

Retrograde Inhibition by a Specific Subset of Interpeduncular Α5 Nicotinic Neurons Regulates Nicotine Preference
Retrograde inhibition by a specific subset of interpeduncular α5 nicotinic neurons regulates nicotine preference Jessica L. Ablesa,b,c, Andreas Görlicha,1, Beatriz Antolin-Fontesa,2,CuidongWanga, Sylvia M. Lipforda, Michael H. Riada, Jing Rend,e,3,FeiHud,e,4,MinminLuod,e,PaulJ.Kennyc, Nathaniel Heintza,f,5, and Ines Ibañez-Tallona,5 aLaboratory of Molecular Biology, The Rockefeller University, New York, NY 10065; bDepartment of Psychiatry, Icahn School of Medicine at Mount Sinai, New York, NY 10029; cDepartment of Neuroscience, Icahn School of Medicine at Mount Sinai, New York, NY 10029; dNational Institute of Biological Sciences, Beijing 102206, China; eSchool of Life Sciences, Tsinghua University, Beijing 100084, China; and fHoward Hughes Medical Institute, The Rockefeller University, New York, NY 10065 Contributed by Nathaniel Heintz, October 23, 2017 (sent for review October 5, 2017; reviewed by Jean-Pierre Changeux and Lorna W. Role) Repeated exposure to drugs of abuse can produce adaptive changes nicotine withdrawal, and optical activation of IPN GABAergic cells that lead to the establishment of dependence. It has been shown that is sufficient to produce a withdrawal syndrome, while blockade of allelic variation in the α5 nicotinic acetylcholine receptor (nAChR) gene GABAergic cells in the IPN reduced symptoms of withdrawal (17). CHRNA5 is associated with higher risk of tobacco dependence. In the Taken together these studies highlight the critical role of α5in brain, α5-containing nAChRs are expressed at very high levels in the regulating behavioral responses to nicotine. Here we characterize two subpopulations of GABAergic interpeduncular nucleus (IPN). Here we identified two nonoverlapping Amigo1 Epyc α + α Amigo1 α Epyc neurons in the IPN that express α5: α5- and α5- neu- 5 cell populations ( 5- and 5- ) in mouse IPN that respond α Amigo1 α Epyc differentially to nicotine.