Genetic Lineage of the Amami Islanders Inferred from Classical Genetic Markers
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Local Dishes Loved by the Nation
Sapporo 1 Hakodate 2 Japan 5 3 Niigata 6 4 Kanazawa 15 7 Sendai Kyoto 17 16 Kobe 10 9 18 20 31 11 8 ocal dishes Hiroshima 32 21 33 28 26 19 13 Fukuoka 34 25 12 35 23 22 14 40 37 27 24 29 Tokyo loved by 41 38 36 Nagoya 42 44 39 30 Shizuoka Yokohama 43 45 Osaka Nagasaki 46 Kochi the nation Kumamoto ■ Hokkaido ■ Tohoku Kagoshima L ■ Kanto ■ Chubu ■ Kansai 47 ■ Chugoku ■ Shikoku Naha ■ Kyushu ■ Okinawa 1 Hokkaido 17 Ishikawa Prefecture 33 Okayama Prefecture 2 Aomori Prefecture 18 Fukui Prefecture 34 Hiroshima Prefecture 3 Iwate Prefecture 19 Yamanashi Prefecture 35 Yamaguchi Prefecture 4 Miyagi Prefecture 20 Nagano Prefecture 36 Tokushima Prefecture 5 Akita Prefecture 21 Gifu Prefecture 37 Kagawa Prefecture 6 Yamagata Prefecture 22 Shizuoka Prefecture 38 Ehime Prefecture 7 Fukushima Prefecture 23 Aichi Prefecture 39 Kochi Prefecture 8 Ibaraki Prefecture 24 Mie Prefecture 40 Fukuoka Prefecture 9 Tochigi Prefecture 25 Shiga Prefecture 41 Saga Prefecture 10 Gunma Prefecture 26 Kyoto Prefecture 42 Nagasaki Prefecture 11 Saitama Prefecture 27 Osaka Prefecture 43 Kumamoto Prefecture 12 Chiba Prefecture 28 Hyogo Prefecture 44 Oita Prefecture 13 Tokyo 29 Nara Prefecture 45 Miyazaki Prefecture 14 Kanagawa Prefecture 30 Wakayama Prefecture 46 Kagoshima Prefecture 15 Niigata Prefecture 31 Tottori Prefecture 47 Okinawa Prefecture 16 Toyama Prefecture 32 Shimane Prefecture Local dishes loved by the nation Hokkaido Map No.1 Northern delights Iwate Map No.3 Cool noodles Hokkaido Rice bowl with Tohoku Uni-ikura-don sea urchin and Morioka Reimen Chilled noodles -

Restoration of the Native Species to Amami Oshima Island
alien species in Amami Oshima Island In addition to the Small Indian mongoose, many other alien species (e.g., feral cats, feral goats, black rats and the Lanceleaf tickseed) have become established on Amami Oshima. Please be sure never to leave behind alien species in the wild nor let them escape. Feral cat Feral goat Black rat Lanceleaf tickseed ● Alien species of Amami Islands HP http://kyushu.env.go.jp/naha/wildlife/data/130902aa.pdf We ask for your cooperation in The mongoose eradication project activity of Amami Mongoose Busters in Amami Oshima The Amami Mongoose Busters, which was formed in 2005, has continued its efforts to eradicate mongooses with the support of people in the island and researchers. We ask for your continued onservation of a precious understanding and support of the mongoose control project as C well as the Amami Mongoose Busters. ecosystem in ■ Amami Mongoose Busters Blog http://amb.amamin.jp/ ■ Amami Mongoose Busters Facebook Amami Oshima Island https://www.facebook.com/amamimongoosebusters March 2014 Amami Wildlife Conservation Center, Published by: Ministry of the Environment, Japan Naha Nature Conservation Office, 551 Koshinohata, Ongachi, Yamato-son, Oshima-gun, Ministry of the Environment, Japan Kagoshima 894-3104 TEL:+81-997-55-8620 Okinawa Tsukansha Building 4F, 5-21 Yamashita-cho, Japan Wildlife Research Center, Naha-shi, Okinawa 900-0027 Amami Ooshima Division (Amami Mongoose Busters) 1385-2 Naze, Uragami,Amami-City, Kagoshima 894-0008 TEL:+81-997-58-4013 Edited by : Japan Wildlife Research Center FOR ALL THE LIFE ON EARTH Design : artpost inc. Photos : Mamoru Tsuneda, Teruho Abe, Yoshihito Goto, Kazuki Yamamuro, Biodiversity Ryuta Yoshihara, Japan Wildlife Research Center Animals and plants Habu snake Protobothrops flavoviridis This poisonous snake is found on Amami Oshima, Tokunoshima, Okinawa in Amami Oshima Island Island, and other several neighboring small islands. -

Mapping Small Islands Communicative Ecologies: a Case Study from the Amami Islands
South Pacific Studies Vol.39, No.1, 2018 Mapping Small Islands Communicative Ecologies: a Case Study from the Amami Islands Papoutsaki Evangelia 1* and Kuwahara Sueo2 1: School of Arts and Science, University of Central Asia, Naryn Campus, 310 Lenin St. Naryn, 722918, Kyrgyz Republic 2: Center for General Education, Kagoshima University *Corresponding author E-mail: [email protected] Abstract This paper provides a conceptual framework for an islands’ communication ecology (CE) approach and an overview of the key findings of exploratory ethnographic research that aimed at mapping the CE of the Amami islands. The communicative ecology approach refers to the various forms, resources, activities, channels and flows of communication and information unique to an island or group of islands, to the island context and identity and a milieu of island agents. This research contributes towards an in-depth understanding of these Islands’ communicative environment; identifies key island mediated communicative networks and practices; explores the role of media in localized information flows unique to the islands; and attempts to explore how small island experiences can help contextualize the theoretical approach of communicative ecology. Key words: Amami Island, Island Media, Island Radio, Islands Communicative Ecology, Mapping Islands Information Ecosystems ―――――――――――――――――――――――――――――――――――――――――――――― Received: 01 September, 2018 Accepted: 30 November, 2018 26 South Pacific Studies Vol.39, No.1, 2018 Introduction This article draws upon a research project aimed at exploring the Amami Islands’ communicative environment by identifying key communicative networks and practices that contribute to sustaining the Amami Islands’ sociocultural cohesion and investigating the role that key mediated forms of communication, particularly community based, play in localized information flows unique to these islands. -

Amami Island Religion - Historical Dynamics of the Islanders’ Spirit - Megumi TAKARABE and Akira NISHIMURA
KAWAI, K., TERADA, R. and KUWAHARA, S. (eds): The Islands of Kagoshima Kagoshima University Research Center for the Pacific Islands, 15 March 2013 Chapter 3 Amami Island Religion - Historical Dynamics of the Islanders’ Spirit - Megumi TAKARABE and Akira NISHIMURA 1. Introduction into “aman’yu” (Amami period),” “aji’yu” (Lords enerally speaking, Japan’s indigenous Shinto period), “nahan’yu” (Ryukyu kingdom period), Gand exogenous Buddhism represent the ma- “yamaton’yu” (Shimadzu controlling period) and jority religions in Japan. These religions have been “america’yu” (American controlling period). Re- recognized as the spiritual pillars of the Japanese. cords are only available from the Naha period on- When compared with the history of religion in Ja- wards and it was the Ryukyu-dominated Amami Is- pan, “Amami Island religion” can be considered lands that welcomed the first unified regime. There unique for its history as well as for its distant loca- are two theories concerning this period, one that it tion from mainland Japan. This is because the reli- began in 1266 (SAKAGUCHI 1921, NOBORI 1949) and gious culture which has existed in various parts of one that it began in 1440 (Richo Jitsuroku). The the Amami Islands comprise a long-standing spiri- latter theory is currently the prevailing view. Ac- tual pillar of the islanders. In other words, the Ama- cordingly, the Amami Islands in the Naha period mi Islands have enjoyed a religious culture of the are said to have lasted for approximately 170 years Ryukyu legacy rather than that of mainland Japan. from 1440 to 1609. This religious culture informs the spiritual base of During the Naha period and the reign of the the Amami Islands today. -

Border-Crossers and Resistance to US Military Rule in the Ryukyus, 1945-1953
Volume 6 | Issue 9 | Article ID 2906 | Sep 01, 2008 The Asia-Pacific Journal | Japan Focus Border-Crossers and Resistance to US Military Rule in the Ryukyus, 1945-1953 Matthew R. Augustine Border-Crossers and Resistance to US main island groups. Trade barriers with Japan Military Rule in the Ryukyus, 1945-1953 were relaxed in 1950, but the San Francisco Peace Treaty signed in 1951 reaffirmed that Matthew R. Augustine the Ryukyus would remain under US military rule, divided from Japan. By this time, the Sixty-six years after Japan’s annexation of the increasing cross-border interconnections former Ryukyu Kingdom in 1879, in the waning between residents in the Ryukyus and months of the Asia-Pacific War, the American Okinawan and Amamian residents in Japan had military partitioned the Ryukyu Islands from already given rise to an organized movement Japan. The replacement of Okinawa Prefecture calling for reversion to Japanese sovereignty. by US military rule in the Ryukyus from 1945 had profound implications, for residents of the How did the establishment of the military occupied islands. A major repercussion of the government and new postwar borders actually military government’s separation of theaffect the movement of residents within and Ryukyus was the enforced isolation of the four outside of the Ryukyu Islands? Conversely, to main island groups from occupied Japan. The what extent did the resistance of residents in Ryukyuan-Japanese border severed long- the Ryukyus and their effort to overcome their standing administrative and economic links, division and isolation influence the military while restrictive border controls prohibited free government’s border controls and related travel and interaction between the two sides. -

The State and Racialization: the Case of Koreans in Japan
The Center for Comparative Immigration Studies CCIS University of California, San Diego The State and Racialization: The Case of Koreans in Japan By Kazuko Suzuki Visiting Fellow, Center for Comparative Immigration Studies Working Paper 69 February 2003 The State and Racialization: The Case of Koreans in Japan Kazuko Suzuki1 Center for Comparative Immigration Studies ********** Abstract. It is frequently acknowledged that the notion of ‘race’ is a socio-political construct that requires constant refurbishment. However, the process and consequences of racialization are less carefully explored. By examining the ideology about nationhood and colonial policies of the Japanese state in relation to Koreans, I will attempt to demonstrate why and how the Japanese state racialized its population. By so doing, I will argue that the state is deeply involved in racialization by fabricating and authorizing ‘differences’ and ‘similarities’ between the dominant and minority groups. Introduction The last decade has seen a growing interest in the state within the field of sociology and political science. While the main contributors of the study have been scholars in comparative and historical sociology and researchers in the economics of development, student of race and ethnicity have gradually paid attention to the role of the state in forming racial/ethnic communities, ethnic identity, and ethnic mobilization (Barkey and Parikh 1991; Marx 1998). State policies clearly constitute one of the major determinants of immigrant adaptation and shifting identity patterns (Hein 1993; Olzak 1983; Nagel 1986). However, the study of the state’s role in race and ethnic studies is still underdeveloped, and many important questions remain to be answered. -

A Guide for the Study of Kinship and Social Organization in the Amami Islands Sumie NAKATANI
KAWAI, K., TERADA, R. and KUWAHARA, S. (eds): The Islands of Kagoshima Kagoshima University Research Center for the Pacific Islands, 15 March 2013 Chapter 9 A Guide for the Study of Kinship and Social Organization in the Amami Islands Sumie NAKATANI 1. Pioneering studies in the 1950s the Tohoku region of northern Japan, where dou- n the 1950s when the Amami Islands were re- zoku, a patrilineal family-kin network was domi- Iturned to Japan, a joint survey was conducted by nant. OYAMA’s study in Yoronjima Is. was finally the members of nine Japanese academic associa- published in the book “Nanseisyoto no Kazoku no tions (Kyu Gakkai Rengo). From 1955 to 1958, a Kenkyu (A Study of Family Systems in the Nansei total of 100 researchers participated in the survey Islands )” (OYAMA 1960). of five of the Amami Islands. Researchers in the In the 1950s, Japanese ethnologists were eager fields of ethnology and sociology were interested to identify the original form or roots of Japanese in kinship relations, locally called hara, haroji, culture. Rural communities of Amami were stud- and hiki, because they seemed to play central roles ied in comparison with those of mainland Japan. in the lives of the people of the Amami Islands. GAMOU, who did his fieldwork in Kikaijima Is. and Several papers based on the survey were focused on the kinship system called haroji, point- published in the journal “Jinrui Kagaku (Human ed out three characteristics of haroji. Science)” and in a book titled “Amami Shizen to 1: The membership of haroji could be traced Bunka (Amami: Nature and Culture)” (OYAMA bilaterally. -

Island Narratives in the Making of Japan: the Kojiki in Geocultural Context
Island Studies Journal, Ahead of print Island narratives in the making of Japan: The Kojiki in geocultural context Henry Johnson University of Otago [email protected] Abstract: Shintō, the national religion of Japan, is grounded in the mythological narratives that are found in the 8th-Century chronicle, Kojiki 古事記 (712). Within this early source book of Japanese history, myth, and national origins, there are many accounts of islands (terrestrial and imaginary), which provide a foundation for comprehending the geographical cosmology (i.e., sacred space) of Japan’s territorial boundaries and the nearby region in the 8th Century, as well as the ritualistic significance of some of the country’s islands to this day. Within a complex geocultural genealogy of gods that links geography to mythology and the Japanese imperial line, land and life were created along with a number of small and large islands. Drawing on theoretical work and case studies that explore the geopolitics of border islands, this article offers a critical study of this ancient work of Japanese history with specific reference to islands and their significance in mapping Japan. Arguing that a characteristic of islandness in Japan has an inherent connection with Shintō religious myth, the article shows how mythological islanding permeates geographic, social, and cultural terrains. The discussion maps the island narratives found in the Kojiki within a framework that identifies and discusses toponymy, geography, and meaning in this island nation’s mythology. Keywords: ancient Japan, border islands, geopolitics, Kojiki, mapping, mythology https://doi.org/10.24043/isj.164 • Received July 2020, accepted April 2021 © Island Studies Journal, 2021 Introduction This study interprets the significance of islands in Japanese mythological history. -

Japanese Reflections on World War II and the American Occupation Japanese Reflections on World War II and the American Occupation Asian History
3 ASIAN HISTORY Porter & Porter and the American Occupation II War World on Reflections Japanese Edgar A. Porter and Ran Ying Porter Japanese Reflections on World War II and the American Occupation Japanese Reflections on World War II and the American Occupation Asian History The aim of the series is to offer a forum for writers of monographs and occasionally anthologies on Asian history. The Asian History series focuses on cultural and historical studies of politics and intellectual ideas and crosscuts the disciplines of history, political science, sociology and cultural studies. Series Editor Hans Hägerdal, Linnaeus University, Sweden Editorial Board Members Roger Greatrex, Lund University Angela Schottenhammer, University of Salzburg Deborah Sutton, Lancaster University David Henley, Leiden University Japanese Reflections on World War II and the American Occupation Edgar A. Porter and Ran Ying Porter Amsterdam University Press Cover illustration: 1938 Propaganda poster “Good Friends in Three Countries” celebrating the Anti-Comintern Pact Cover design: Coördesign, Leiden Lay-out: Crius Group, Hulshout Amsterdam University Press English-language titles are distributed in the US and Canada by the University of Chicago Press. isbn 978 94 6298 259 8 e-isbn 978 90 4853 263 6 doi 10.5117/9789462982598 nur 692 © Edgar A. Porter & Ran Ying Porter / Amsterdam University Press B.V., Amsterdam 2017 All rights reserved. Without limiting the rights under copyright reserved above, no part of this book may be reproduced, stored in or introduced into a retrieval system, or transmitted, in any form or by any means (electronic, mechanical, photocopying, recording or otherwise) without the written permission of both the copyright owner and the author of the book. -

Those on the Latter by Katayama (I) and Minamibuchi (I)
GENETIC STRUCTURE OF HUMAN POPULATIONS III. DIFFERENTIATION OF ABO BLOOD GROUP GENE FREQUENCIES IN SMALL AREAS OF JAPAN* MASATOSHI NEI and YOKO IMAIZUMI Division of Genetics, National Institute of Radiological Sciences, Chiba, Japan Receivedii .xii.65 1.INTRODUCTION INa previous paper (Nei and Imaizumi, 1966a) it was shown that the local differentiation of ABO and MitT blood group gene frequencies in Japan has occurred largely by genetic random drift. In that investi- gation the population of Japan was divided into 45 different sub- populations or Prefectures (administrative units of Japan) the sizes of which were mostly one to two millions. However, the size of mating groups of neighbourhoods in Wright's (1946) sense appears to be much smaller than the sub-populations employed in the paper. It is, there- fore, expected that the gene frequencies are locally differentiated even within these sub-populations. We thus examined the degree of differ- entiation of ABO blood group gene frequencies in several small areas of Japan. 2.DIFFERENTIATION WITHIN PREFECTURES Thereare two Prefectures, in which the local variation of ABO blood group gene frequencies can be analysed. One is Kagoshima Prefecture in Kyushu Island and the other Tokushima Prefecture in Shikoku Island (cf fig. i in Nei and Imaizumi, 1966a). The data on the former were collected by Ono and Takagi (1942) and Makisumi (1958), and those on the latter by Katayama (i) and Minamibuchi (i). The areas of Kagoshima and Tokushima Prefectures are 9,104 km2 and 4,143 km2 respectively, both including several neighbouring small islands. The population sizes of these two Prefectures at the time of 1960 census are 1,962,998 and 847,279 respectively. -

Human and Physical Geography of Japan Study Tour 2012 Reports
Five College Center for East Asian Studies National Consortium for Teaching about Asia (NCTA) 2012 Japan Study Tour The Human and Physical Geography of Japan Reports from the Field United States Department of Education Fulbright-Hays Group Project Abroad with additional funding from the Freeman Foundation Five College Center for East Asian Studies 69 Paradise Road, Florence Gilman Pavilion Northampton, MA 01063 The Human and Physical Geography of Japan Reports from the Field In the summer of 2012, twelve educators from across the United States embarked on a four-week journey to Japan with the goal of enriching their classroom curriculum content by learning first-hand about the country. Prior to applying for the study tour, each participant completed a 30-hour National Consortium for Teaching about Asia (NCTA) seminar. Once selected, they all completed an additional 20 hours of pre-departure orientation, including FCCEAS webinars (funded by the US-Japan Foundation; archived webinars are available at www.smith.edu/fcceas), readings, and language podcasts. Under the overarching theme of “Human and Physical Geography of Japan,” the participants’ experience began in Tokyo, then continued in Sapporo, Yokohama, Kamakura, Kyoto, Osaka, Nara, Hiroshima, Miyajima, and finally ended in Naha. Along the way they heard from experts on Ainu culture and burakumin, visited the Tokyo National Museum of History, heard the moving testimony of an A-bomb survivor, toured the restored seat of the Ryukyu Kingdom, and dined on regional delicacies. Each study tour participant was asked to prepare a report on an assigned geography-related topic to be delivered to the group in country and then revised upon their return to the U.S. -
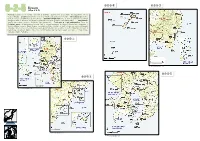
6-2-5-④ 6-2-5-③ Kyusyu 6-2-5 (Map 6-2-5) Province: Kumamoto Pref
6-2-5-④ 6-2-5-③ Kyusyu 6-2-5 (Map 6-2-5) Province: Kumamoto Pref. at west, Oita Pref. at northeast, Miyazaki Pref. at southeast, and Kagoshima Pref. at south of Kyushu Location: Kyushu lies at west of Shikoku and southwest of Honshu Air temperature: 17.8˚C (annual average, at Ushibuka City, Kumamoto.) Seawater temperature: 22.9˚C, 22.2 ˚C and 20.9 ˚C (annual average, at east off Aburatsu, southwest off Kushikijima (Is.) and Yatsushiro Sea, respectively) Precipitation: 2,027.9 mm (annual average, Ushibuka City, Kumamoto) Total area of coral communities: 581.8 ha Protected areas: Unzen-Amakusa National Park: at around Amakusa, including 3 Marine Park Zones and 2 Protected Water Surfaces; Nippo Kaigan Quasi-National Park: coastline at south of Oita and north of Miyazaki, including 2 Marine Park Zones; Nichinan Kaigan Quasi-National Park: coastline at south of Miyazaki and east of Kagoshima, including 1 Marine Park Zone; Kirishima-Yaku National Park: a part of coastline in Kagoshima, including 2 Marine Park Zones. 6-2-5-① *“号”on this map means“site”. 6-2-5-⑤ 6-2-5-② *“号”on this map means“site”. 6-2-5-② 6-2-5-① *“号”on this map means“site”. 6-2-5-④ 6-2-5-③ *“号”on this map means“site”. 6-2-5-⑤ 06 Coral Reefs of Japan a. Kumamoto Prefecture (Map 6-2-5-①) Satoshi Nojima 1 Corals and coral reefs Photo. 1. Tabulate Acropora dominant community in Kuwashima 1. Geographical features (Is.), Ushibuka City, Kumamoto Prefecture. On the west of Kyushu in Kumamoto Prefecture lie the Amakusa Islands.