Knockout of STK10 Promotes the Migration and Invasion of Cervical Cancer Cells
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Drug Sensitivity and Resistance Testing (DSRT) Approach
A phenotypic screening and machine learning platform eciently identifies triple negative breast cancer-selective and readily druggable targets Prson Gautam 1 Alok Jaiswal 1 Tero Aittokallio 1, 2 Hassan Al Ali 3 Krister Wennerberg 1,4 Identifying eective oncogenic targets is challenged by the complexity of genetic alterations in 1Institute for Molecular Medicine Finland (FIMM), HiLIFE, University of Helsinki, Finland cancer and their poorly understood relation to cell function and survival. There is a need for meth- Current kinome coverage of kinase inhibitors in TNBC exhibit diverse kinase dependencies MFM-223 is selectively addicted to FGFR2 2Department of Mathematics and Statistics, University of Turku, Finland 3The Miami Project to Cure Paralysis, Peggy and Harold Katz Family Drug Discovery Center, A A Sylvester Comprehensive Cancer Center, and Department of Neurological Surgery and Medicine ods that rapidly and accurately identify “pharmacologically eective” targets without the require- clinical evaluation TN Kinases MFM-223 CAL-120 MDA-MB-231 TNBC TNBC TNBC TNBC TNBC TNBC HER2+ 100 University of Miami Miller School of Medicine, Miami, FL 33136, USA. non- HER2+ FGFR1 0.97 0.00 0.00 MFM-223 BL1 BL2 M MSL IM LAR ER+, PR+ 50 ment for priori knowledge of complex signaling networks. We developed an approach that uses ma- cancerous FGFR2 56.46 0.00 0.00 CAL-120 25 4 MDA-MB-231 Biotech Research & Innovation Centre (BRIC) and Novo Nordisk Foundation Center HCC1937 CAL-85-1 CAL-120 MDA-MB-231 DU4475 CAL-148 MCF-10A SK-BR-3 BT-474 FGFR3 25.10 0.00 0.00 0 chine learning to relate results from unbiased phenotypic screening of kinase inhibitors to their bio- for Stem Cell Biology (DanStem), University of Copenhagen, Denmark HCC1599 HDQ-P1 BT-549 MDA-MB-436 MFM-223 FGFR4 0.00 0.00 0.00 MAXIS*Bk Clinical status MDA-MB-468 CAL-51 Hs578T MDA-MB-453 score chemical activity data. -

Clinical Utility of Recently Identified Diagnostic, Prognostic, And
Modern Pathology (2017) 30, 1338–1366 1338 © 2017 USCAP, Inc All rights reserved 0893-3952/17 $32.00 Clinical utility of recently identified diagnostic, prognostic, and predictive molecular biomarkers in mature B-cell neoplasms Arantza Onaindia1, L Jeffrey Medeiros2 and Keyur P Patel2 1Instituto de Investigacion Marques de Valdecilla (IDIVAL)/Hospital Universitario Marques de Valdecilla, Santander, Spain and 2Department of Hematopathology, MD Anderson Cancer Center, Houston, TX, USA Genomic profiling studies have provided new insights into the pathogenesis of mature B-cell neoplasms and have identified markers with prognostic impact. Recurrent mutations in tumor-suppressor genes (TP53, BIRC3, ATM), and common signaling pathways, such as the B-cell receptor (CD79A, CD79B, CARD11, TCF3, ID3), Toll- like receptor (MYD88), NOTCH (NOTCH1/2), nuclear factor-κB, and mitogen activated kinase signaling, have been identified in B-cell neoplasms. Chronic lymphocytic leukemia/small lymphocytic lymphoma, diffuse large B-cell lymphoma, follicular lymphoma, mantle cell lymphoma, Burkitt lymphoma, Waldenström macroglobulinemia, hairy cell leukemia, and marginal zone lymphomas of splenic, nodal, and extranodal types represent examples of B-cell neoplasms in which novel molecular biomarkers have been discovered in recent years. In addition, ongoing retrospective correlative and prospective outcome studies have resulted in an enhanced understanding of the clinical utility of novel biomarkers. This progress is reflected in the 2016 update of the World Health Organization classification of lymphoid neoplasms, which lists as many as 41 mature B-cell neoplasms (including provisional categories). Consequently, molecular genetic studies are increasingly being applied for the clinical workup of many of these neoplasms. In this review, we focus on the diagnostic, prognostic, and/or therapeutic utility of molecular biomarkers in mature B-cell neoplasms. -

Phosphorylation Sites in Protein Kinases and Phosphatases Regulated by Formyl Peptide Receptor 2 Signaling
International Journal of Molecular Sciences Review Phosphorylation Sites in Protein Kinases and Phosphatases Regulated by Formyl Peptide Receptor 2 Signaling Maria Carmela Annunziata 1, Melania Parisi 1, Gabriella Esposito 2 , Gabriella Fabbrocini 1, Rosario Ammendola 2 and Fabio Cattaneo 2,* 1 Department of Clinical Medicine and Surgery, School of Medicine, University of Naples Federico II, Via S. Pansini 5, 80131 Naples, Italy; [email protected] (M.C.A.); [email protected] (M.P.); [email protected] (G.F.) 2 Department of Molecular Medicine and Medical Biotechnology, School of Medicine,, University of Naples Federico II, Via S. Pansini 5, 80131 Naples, Italy; [email protected] (G.E.); [email protected] (R.A.) * Correspondence: [email protected]; Fax: +39-081-7464-359 Received: 5 May 2020; Accepted: 25 May 2020; Published: 27 May 2020 Abstract: FPR1, FPR2, and FPR3 are members of Formyl Peptides Receptors (FPRs) family belonging to the GPCR superfamily. FPR2 is a low affinity receptor for formyl peptides and it is considered the most promiscuous member of this family. Intracellular signaling cascades triggered by FPRs include the activation of different protein kinases and phosphatase, as well as tyrosine kinase receptors transactivation. Protein kinases and phosphatases act coordinately and any impairment of their activation or regulation represents one of the most common causes of several human diseases. Several phospho-sites has been identified in protein kinases and phosphatases, whose role may be to expand the repertoire of molecular mechanisms of regulation or may be necessary for fine-tuning of switch properties. We previously performed a phospho-proteomic analysis in FPR2-stimulated cells that revealed, among other things, not yet identified phospho-sites on six protein kinases and one protein phosphatase. -

Comparative Genomic Evidence for Self-Domestication in Homo Sapiens
bioRxiv preprint doi: https://doi.org/10.1101/125799; this version posted April 9, 2017. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Comparative genomic evidence for self-domestication in Homo sapiens Constantina Theofanopoulou1,2,a, Simone Gastaldon1,a, Thomas O’Rourke1, Bridget D. Samuels3, Angela Messner1, Pedro Tiago Martins1, Francesco Delogu4, Saleh Alamri1, and Cedric Boeckx1,2,5,b 1Section of General Linguistics, Universitat de Barcelona 2Universitat de Barcelona Institute for Complex Systems 3Center for Craniofacial Molecular Biology, University of Southern California 4Department of Chemistry, Biotechnology and Food Science, Norwegian University of Life Sciences (NMBU) 5ICREA aThese authors contributed equally to this work bCorresponding author: [email protected] ABSTRACT This study identifies and analyzes statistically significant overlaps between selective sweep screens in anatomically modern humans and several domesticated species. The results obtained suggest that (paleo-)genomic data can be exploited to complement the fossil record and support the idea of self-domestication in Homo sapiens, a process that likely intensified as our species populated its niche. Our analysis lends support to attempts to capture the “domestication syndrome” in terms of alterations to certain signaling pathways and cell lineages, such as the neural crest. Introduction Recent advances in genomics, coupled with other sources of information, offer new opportunities to test long-standing hypotheses about human evolution. Especially in the domain of cognition, the retrieval of ancient DNA could, with the help of well-articulated linking hypotheses connecting genes, brain and cognition, shed light on the emergence of ‘cognitive modernity’. -

Clinical Potential of Kinase Inhibitors in Combination with Immune Checkpoint Inhibitors for the Treatment of Solid Tumors
International Journal of Molecular Sciences Review Clinical Potential of Kinase Inhibitors in Combination with Immune Checkpoint Inhibitors for the Treatment of Solid Tumors Ryuhjin Ahn 1 and Josie Ursini-Siegel 2,3,4,5,* 1 Department of Biological Engineering, Koch Institute for Integrative Cancer Research, Massachusetts Institute of Technology, Cambridge, MA 02139, USA; [email protected] 2 Department of Biochemistry, McGill University, Montréal, QC H3G 1Y6, Canada 3 Lady Davis Institute for Medical Research, Jewish General Hospital, Montréal, QC H3T 1E2, Canada 4 Department of Experimental Medicine, McGill University, Montréal, QC H3A 0G4, Canada 5 Department of Oncology, McGill University, 546 Pine Avenue West, Montréal, QC H2W 1S6, Canada * Correspondence: [email protected]; Tel.: +514-340-8222 (ext. 26557); Fax: +514-340-7502 Abstract: Oncogenic kinases contribute to immunosuppression and modulate the tumor microenvi- ronment in solid tumors. Increasing evidence supports the fundamental role of oncogenic kinase signaling networks in coordinating immunosuppressive tumor microenvironments. This has led to numerous studies examining the efficacy of kinase inhibitors in inducing anti-tumor immune responses by increasing tumor immunogenicity. Kinase inhibitors are the second most common FDA-approved group of drugs that are deployed for cancer treatment. With few exceptions, they inevitably lead to intrinsic and/or acquired resistance, particularly in patients with metastatic disease when used as a monotherapy. On the other hand, cancer immunotherapies, including immune checkpoint inhibitors, have revolutionized cancer treatment for malignancies such as melanoma and Citation: Ahn, R.; Ursini-Siegel, J. lung cancer. However, key hurdles remain to successfully incorporate such therapies in the treatment Clinical Potential of Kinase Inhibitors of other solid cancers. -

Activation of Diverse Signalling Pathways by Oncogenic PIK3CA Mutations
ARTICLE Received 14 Feb 2014 | Accepted 12 Aug 2014 | Published 23 Sep 2014 DOI: 10.1038/ncomms5961 Activation of diverse signalling pathways by oncogenic PIK3CA mutations Xinyan Wu1, Santosh Renuse2,3, Nandini A. Sahasrabuddhe2,4, Muhammad Saddiq Zahari1, Raghothama Chaerkady1, Min-Sik Kim1, Raja S. Nirujogi2, Morassa Mohseni1, Praveen Kumar2,4, Rajesh Raju2, Jun Zhong1, Jian Yang5, Johnathan Neiswinger6, Jun-Seop Jeong6, Robert Newman6, Maureen A. Powers7, Babu Lal Somani2, Edward Gabrielson8, Saraswati Sukumar9, Vered Stearns9, Jiang Qian10, Heng Zhu6, Bert Vogelstein5, Ben Ho Park9 & Akhilesh Pandey1,8,9 The PIK3CA gene is frequently mutated in human cancers. Here we carry out a SILAC-based quantitative phosphoproteomic analysis using isogenic knockin cell lines containing ‘driver’ oncogenic mutations of PIK3CA to dissect the signalling mechanisms responsible for oncogenic phenotypes induced by mutant PIK3CA. From 8,075 unique phosphopeptides identified, we observe that aberrant activation of PI3K pathway leads to increased phosphorylation of a surprisingly wide variety of kinases and downstream signalling networks. Here, by integrating phosphoproteomic data with human protein microarray-based AKT1 kinase assays, we discover and validate six novel AKT1 substrates, including cortactin. Through mutagenesis studies, we demonstrate that phosphorylation of cortactin by AKT1 is important for mutant PI3K-enhanced cell migration and invasion. Our study describes a quantitative and global approach for identifying mutation-specific signalling events and for discovering novel signalling molecules as readouts of pathway activation or potential therapeutic targets. 1 McKusick-Nathans Institute of Genetic Medicine and Department of Biological Chemistry, Johns Hopkins University School of Medicine, 733 North Broadway, BRB 527, Baltimore, Maryland 21205, USA. -
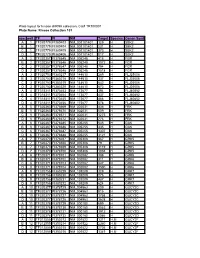
Kinase Collection 101 Row Well TF FI NM Target Location Spec
Plate layout for kinase shRNA collection, Cat# TR100001 Plate Name: Kinase Collection 101 row well TF FI NM Target LocationSpecies Genen Symbol A 1 TF320779 FI380403 NM_001101401 129 H SBK2 B 1 TF320779 FI380404 NM_001101401 231 H SBK2 C 1 TF320779 FI380405 NM_001101401 373 H SBK2 D 1 TF320779 FI380406 NM_001101401 817 H SBK2 A 2 TF320357 FI378645 NM_005248 415 H FGR B 2 TF320357 FI378646 NM_005248 1072 H FGR C 2 TF320357 FI378647 NM_005248 794 H FGR D 2 TF320357 FI378648 NM_005248 1518 H FGR A 3 TF320753 FI380217 NM_144610 289 H FLJ25006 B 3 TF320753 FI380218 NM_144610 137 H FLJ25006 C 3 TF320753 FI380219 NM_144610 642 H FLJ25006 D 3 TF320753 FI380220 NM_144610 673 H FLJ25006 A 4 TF318311 FI370453 NM_173677 394 H FLJ40852 B 4 TF318311 FI370454 NM_173677 437 H FLJ40852 C 4 TF318311 FI370455 NM_173677 466 H FLJ40852 D 4 TF318311 FI370456 NM_173677 578 H FLJ40852 A 5 TF320363 FI378669 NM_002031 436 H FRK B 5 TF320363 FI378670 NM_002031 509 H FRK C 5 TF320363 FI378671 NM_002031 1275 H FRK D 5 TF320363 FI378672 NM_002031 776 H FRK A 6 TF320367 FI378685 NM_005255 545 H GAK B 6 TF320367 FI378686 NM_005255 435 H GAK C 6 TF320367 FI378687 NM_005255 1307 H GAK D 6 TF320367 FI378688 NM_005255 3121 H GAK A 7 TF320370 FI378697 NM_005308 367 H GRK5 B 7 TF320370 FI378698 NM_005308 79 H GRK5 C 7 TF320370 FI378699 NM_005308 1119 H GRK5 D 7 TF320370 FI378700 NM_005308 1038 H GRK5 A 8 TF320371 FI378701 NM_002082 388 H GRK6 B 8 TF320371 FI378702 NM_002082 311 H GRK6 C 8 TF320371 FI378703 NM_002082 847 H GRK6 D 8 TF320371 FI378704 NM_002082 1590 H GRK6 A -

STK10 Knockout Inhibits Cell Migration and Promotes Cell Proliferation Via Modulating the Activity of ERM and P38 MAPK in Prostate Cancer Cells
EXPERIMENTAL AND THERAPEUTIC MEDICINE 22: 851, 2021 STK10 knockout inhibits cell migration and promotes cell proliferation via modulating the activity of ERM and p38 MAPK in prostate cancer cells LU ZHANG1, SHUN‑YUAN LU1, RUI GUO1, JIN‑XIA MA1, LING‑YUN TANG1, JIN‑JIN WANG2, CHUN‑LING SHEN1, LI‑MING LU3, JIE LIU4, ZHU‑GANG WANG1 and HONG‑XIN ZHANG1 1Research Center for Experimental Medicine, State Key Laboratory of Medical Genomics, Shanghai Ruijin Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai 200025; 2Shanghai Model Organisms Center, Shanghai 201321; 3Shanghai Institute of Immunology, Shanghai Jiao Tong University School of Medicine; 4Shanghai Institute of Orthopaedics and Traumatology, Shanghai Ruijin Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai 200025, P.R. China Received March 9, 2020; Accepted May 25, 2021 DOI: 10.3892/etm.2021.10283 Abstract. Prostate cancer (PCa) is one of the most common Introduction types of cancer and is a serious threat to men's health due to the high rate of incidence and metastasis. However, the exact Prostate cancer (PCa) is one of the most common types of underlying pathology of this malignant disease has yet to be malignant cancer affecting the urinary system in men, with fully elucidated. The ezrin‑radixin‑moesin (ERM) family of >1 million new cases diagnosed every year (1). It is a major proteins are associated with the development and metastasis of leading cause of cancer‑related mortality, a serious threat to various types of cancer. Serine threonine kinase 10 (STK10) men's health and imposes a heavy economic burden world‑ is an ERM kinase that is involved in the activation of ERM wide. -

Human STK10 / LOK Protein (His Tag)
Human STK10 / LOK Protein (His Tag) Catalog Number: 11724-H07E General Information SDS-PAGE: Gene Name Synonym: LOK; PRO2729 Protein Construction: A DNA sequence encoding the human STK10 (NP_005981.3) (Arg 18-Glu 317) was expressed, with a polyhistide tag at the N-terminus. Source: Human Expression Host: E. coli QC Testing Purity: > 95 % as determined by SDS-PAGE Bio Activity: Protein Description The specific activity was determined to be 1353 nmol/min/mg using synthetic AXLtide peptide (KKSRGDYMTMQIG) as substrate. Serine / threonine-protein kinase 10, also known as Lymphocyte-oriented kinase, STK10 and LOK, which belongs to theprotein kinase superfamily, Endotoxin: STE Ser / Thr protein kinase family and STE20 subfamily. Protein kinases constitute a large superfamily of enzymes with key regulatory functions in Please contact us for more information. nearly all signal transmission processes of eukaryotic cells. The Ste20 family of serine/threonine kinases plays an important role in numerous Stability: cellular functions such as growth, apoptosis, and morphogenesis. STK10 is Samples are stable for up to twelve months from date of receipt at -70 ℃ similar to several known polo-like kinase kinases. It can associate with and phosphorylate polo-like kinase 1, and overexpression of a kinase-dead Predicted N terminal: Met version of the protein interferes with normal cell cycle progression. STK10 can also negatively regulate interleukin 2 expression in T-cells via the Molecular Mass: mitogen activated protein kinase kinase 1 pathway. Stk10 can associate with Plk1 in cells and furthermore can phosphorylate Plk1. It can also act The recombinant human STK10 consisting of 315 amino acids and has a on substrates such as myelin basic protein and histone 2A on serine and calculated molecular mass of 36 kDa. -

The Greatwall Kinase Safeguards the Genome Integrity by Affecting the Kinome Activity in Mitosis
Oncogene (2020) 39:6816–6840 https://doi.org/10.1038/s41388-020-01470-1 ARTICLE The Greatwall kinase safeguards the genome integrity by affecting the kinome activity in mitosis 1,9 1 2 1,10 1,3 1 Xavier Bisteau ● Joann Lee ● Vinayaka Srinivas ● Joanna H. S. Lee ● Joanna Niska-Blakie ● Gifford Tan ● 1 1 4 5 1,6 7 Shannon Y. X. Yap ● Kevin W. Hom ● Cheng Kit Wong ● Jeongjun Chae ● Loo Chien Wang ● Jinho Kim ● 4 1,6 1,3,11 1,8 Giulia Rancati ● Radoslaw M. Sobota ● Chris S. H. Tan ● Philipp Kaldis Received: 17 June 2020 / Revised: 21 August 2020 / Accepted: 10 September 2020 / Published online: 25 September 2020 © The Author(s) 2020. This article is published with open access Abstract Progression through mitosis is balanced by the timely regulation of phosphorylation and dephosphorylation events ensuring the correct segregation of chromosomes before cytokinesis. This balance is regulated by the opposing actions of CDK1 and PP2A, as well as the Greatwall kinase/MASTL. MASTL is commonly overexpressed in cancer, which makes it a potential therapeutic anticancer target. Loss of Mastl induces multiple chromosomal errors that lead to the accumulation of micronuclei and multilobulated cells in mitosis. Our analyses revealed that loss of Mastl leads to chromosome breaks and abnormalities 1234567890();,: 1234567890();,: impairing correct segregation. Phospho-proteomic data for Mastl knockout cells revealed alterations in proteins implicated in multiple processes during mitosis including double-strand DNA damage repair. In silico prediction of the kinases with affected activity unveiled NEK2 to be regulated in the absence of Mastl. We uncovered that, RAD51AP1, involved in regulation of homologous recombination, is phosphorylated by NEK2 and CDK1 but also efficiently dephosphorylated by PP2A/B55. -
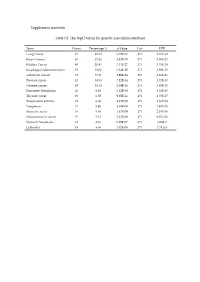
Supplement Materials Table S1. the Top15 Terms for Genetic Association
Supplement materials Table S1. The top15 terms for genetic association database. Term Count Percentage % p-Value List FDR Lung Cancer 73 25.34 6.70E-36 271 1.16E-32 Breast Cancer 67 23.26 2.92E-30 271 5.06E-27 Bladder Cancer 60 20.83 3.33E-27 271 5.78E-24 Esophageal adenocarcinoma 52 18.06 6.24E-29 271 1.08E-25 Colorectal Cancer 51 17.71 4.99E-24 271 8.66E-21 Prostate cancer 42 14.58 7.12E-14 271 1.23E-10 Ovarian cancer 39 13.54 2.04E-16 271 3.89E-13 Pancreatic Neoplasms 20 6.94 1.45E-08 271 2.52E-05 Thyroid cancer 19 6.59 9.98E-11 271 1.73E-07 Rheumatoid arthritis 19 6.60 8.82E-08 271 1.53E-04 Neoplasms 17 5.90 4.58E-08 271 7.94E-05 Stomach cancer 16 5.56 1.17E-09 271 2.03E-06 Head and neck cancer 15 5.21 5.23E-09 271 9.07E-06 Stomach Neoplasms 14 4.86 6.20E-07 271 1.08E-3 Leukemia 14 4.86 1.02E-06 271 1.78 E-3 Table S2. The results of the reverse docking for AutoDock Vina. PDB Protein Gene_Name Uniprot_ID Score 4TVJ Poly [ADP-ribose] polymerase 2 PARP2 Q9UGN5 -10.80 2DQ7 Proto-oncogene tyrosine-protein kinase Fyn FYN P06241 -10.00 4KIK Inhibitor of nuclear factor kappa-B kinase IKBKB O14920 -10.00 subunit beta 3EQR Activated CDC42 kinase 1 ACK1/TNK2 Q07912 -9.90 3MTF Activin receptor type-1 ACVR1 Q04771 -9.90 4GV0 Poly [ADP-ribose] polymerase 3 PARP3 Q9Y6F1 -9.90 1M6I Programmed cell death protein 8 AIF O95831 -9.80 1UA2 Cell division protein kinase 7 CDK7 P50613 -9.80 1Z6T Apoptotic protease activating factor 1 APAF1 O14727 -9.80 1ZXM DNA topoisomerase II TOPII P11388 -9.80 2WOU Serine/Threonine-protein kinase NEK7 NEK7 Q8TDX7 -9.60 -

1 Novel Expression Signatures Identified by Transcriptional Analysis
ARD Online First, published on October 8, 2009 as 10.1136/ard.2009.108043 Ann Rheum Dis: first published as 10.1136/ard.2009.108043 on 7 October 2009. Downloaded from Novel expression signatures identified by transcriptional analysis of separated leukocyte subsets in SLE and vasculitis 1Paul A Lyons, 1Eoin F McKinney, 1Tim F Rayner, 1Alexander Hatton, 1Hayley B Woffendin, 1Maria Koukoulaki, 2Thomas C Freeman, 1David RW Jayne, 1Afzal N Chaudhry, and 1Kenneth GC Smith. 1Cambridge Institute for Medical Research and Department of Medicine, Addenbrooke’s Hospital, Hills Road, Cambridge, CB2 0XY, UK 2Roslin Institute, University of Edinburgh, Roslin, Midlothian, EH25 9PS, UK Correspondence should be addressed to Dr Paul Lyons or Prof Kenneth Smith, Department of Medicine, Cambridge Institute for Medical Research, Addenbrooke’s Hospital, Hills Road, Cambridge, CB2 0XY, UK. Telephone: +44 1223 762642, Fax: +44 1223 762640, E-mail: [email protected] or [email protected] Key words: Gene expression, autoimmune disease, SLE, vasculitis Word count: 2,906 The Corresponding Author has the right to grant on behalf of all authors and does grant on behalf of all authors, an exclusive licence (or non-exclusive for government employees) on a worldwide basis to the BMJ Publishing Group Ltd and its Licensees to permit this article (if accepted) to be published in Annals of the Rheumatic Diseases and any other BMJPGL products to exploit all subsidiary rights, as set out in their licence (http://ard.bmj.com/ifora/licence.pdf). http://ard.bmj.com/ on October 2, 2021 by guest. Protected copyright. 1 Copyright Article author (or their employer) 2009.