Detection of Cryptosporidium Parvum in a Red-Eared Slider Turtle (Trachemys Scripta Elegans), a Noted Invasive Alien Species, Ca
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Case of Deirocheline Turtles
bioRxiv preprint doi: https://doi.org/10.1101/556670; this version posted February 21, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 1 Body coloration and mechanisms of colour production in Archelosauria: 2 The case of deirocheline turtles 3 Jindřich Brejcha1,2*†, José Vicente Bataller3, Zuzana Bosáková4, Jan Geryk5, 4 Martina Havlíková4, Karel Kleisner1, Petr Maršík6, Enrique Font7 5 1 Department of Philosophy and History of Science, Faculty of Science, Charles University, Viničná 7, Prague 6 2, 128 00, Czech Republic 7 2 Department of Zoology, Natural History Museum, National Museum, Václavské nám. 68, Prague 1, 110 00, 8 Czech Republic 9 3 Centro de Conservación de Especies Dulceacuícolas de la Comunidad Valenciana. VAERSA-Generalitat 10 Valenciana, El Palmar, València, 46012, Spain. 11 4 Department of Analytical Chemistry, Faculty of Science, Charles University, Hlavova 8, Prague 2, 128 43, 12 Czech Republic 13 5 Department of Biology and Medical Genetics, 2nd Faculty of Medicine, Charles University and University 14 Hospital Motol, V Úvalu 84, 150 06 Prague, Czech Republic 15 6 Department of Food Science, Faculty of Agrobiology, Food, and Natural Resources, Czech University of Life 16 Sciences, Kamýcká 129, Prague 6, 165 00, Czech Republic 17 7 Ethology Lab, Cavanilles Institute of Biodiversity and Evolutionary Biology, University of Valencia, C/ 18 Catedrátic José Beltrán Martinez 2, Paterna, València, 46980, Spain 19 Keywords: Chelonia, Trachemys scripta, Pseudemys concinna, nanostructure, pigments, chromatophores 20 21 Abstract 22 Animal body coloration is a complex trait resulting from the interplay of multiple colour-producing mechanisms. -
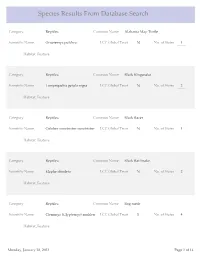
Species Results from Database Search
Species Results From Database Search Category Reptiles Common Name Alabama Map Turtle Scientific Name Graptemys pulchra LCC Global Trust N No. of States 1 Habitat_Feature Category Reptiles Common Name Black Kingsnake Scientific Name Lampropeltis getula nigra LCC Global Trust N No. of States 2 Habitat_Feature Category Reptiles Common Name Black Racer Scientific Name Coluber constrictor constrictor LCC Global Trust N No. of States 1 Habitat_Feature Category Reptiles Common Name Black Rat Snake Scientific Name Elaphe obsoleta LCC Global Trust N No. of States 2 Habitat_Feature Category Reptiles Common Name Bog turtle Scientific Name Clemmys (Glyptemys) muhlen LCC Global Trust Y No. of States 4 Habitat_Feature Monday, January 28, 2013 Page 1 of 14 Category Reptiles Common Name Broadhead Skink Scientific Name Eumeces laticeps LCC Global Trust N No. of States 5 Habitat_Feature Category Reptiles Common Name Coal Skink Scientific Name Eumeces anthracinus LCC Global Trust Y No. of States 8 Habitat_Feature Category Reptiles Common Name Common Five-lined Skink Scientific Name Eumeces fasciatus LCC Global Trust N No. of States 2 Habitat_Feature Category Reptiles Common Name Common Map Turtle Scientific Name Graptemys geographica LCC Global Trust N No. of States 6 Habitat_Feature Category Reptiles Common Name Common Musk Turtle Scientific Name Sternotherus odoratus LCC Global Trust N No. of States 2 Habitat_Feature Monday, January 28, 2013 Page 2 of 14 Category Reptiles Common Name Common Ribbonsnake Scientific Name Thamnophis sauritus sauritus LCC Global Trust N No. of States 6 Habitat_Feature Category Reptiles Common Name Common Snapping Turtle Scientific Name Chelydra serpentina LCC Global Trust N No. of States 2 Habitat_Feature Category Reptiles Common Name Corn snake Scientific Name Elaphe guttata guttata LCC Global Trust N No. -
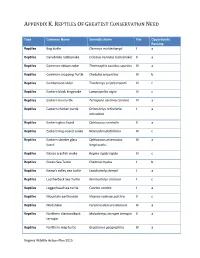
Reptiles of Greatest Conservation Need
APPENDIX K. REPTILES OF GREATEST CONSERVATION NEED Taxa Common Name Scientific Name Tier Opportunity Ranking Reptiles Bog turtle Clemmys muhlenbergii I a Reptiles Canebrake rattlesnake Crotalus horridus (canebrake) II a Reptiles Common ribbonsnake Thamnophis sauritus sauritus IV a Reptiles Common snapping Turtle Chelydra serpentina IV b Reptiles Cumberland slider Trachemys scripta troostii III c Reptiles Eastern black kingsnake Lampropeltis nigra III c Reptiles Eastern box turtle Terrapene carolina carolina III a Reptiles Eastern chicken turtle Deirochelys reticularia I a reticularia Reptiles Eastern glass lizard Ophisaurus ventralis II a Reptiles Eastern hog-nosed snake Heterodon platirhinos IV c Reptiles Eastern slender glass Ophisaurus attenuatus IV a lizard longicaudus Reptiles Glossy crayfish snake Regina rigida rigida III c Reptiles Green Sea Turtle Chelonia mydas I b Reptiles Kemp's ridley sea turtle Lepidochelys kempii I a Reptiles Leatherback Sea Turtle Dermochelys coriacea I c Reptiles Loggerhead sea turtle Caretta caretta I a Reptiles Mountain earthsnake Virginia valeriae pulchra II c Reptiles Mudsnake Farancia abacura abacura IV a Reptiles Northern diamondback Malaclemys terrapin terrapin II a terrapin Reptiles Northern map turtle Graptemys geographica IV a Virginia Wildlife Action Plan 2015 APPENDIX K. REPTILES OF GREATEST CONSERVATION NEED Reptiles Northern pinesnake Pituophis melanoleucus I a melanoleucus Reptiles Queen snake Regina septemvittata IV a Reptiles Rainbow snake Farancia erytrogramma IV a erytrogramma Reptiles -

In AR, FL, GA, IA, KY, LA, MO, OH, OK, SC, TN, and TX): Species in Red = Depleted to the Point They May Warrant Federal Endangered Species Act Listing
Southern and Midwestern Turtle Species Affected by Commercial Harvest (in AR, FL, GA, IA, KY, LA, MO, OH, OK, SC, TN, and TX): species in red = depleted to the point they may warrant federal Endangered Species Act listing Common snapping turtle (Chelydra serpentina) – AR, GA, IA, KY, MO, OH, OK, SC, TX Florida common snapping turtle (Chelydra serpentina osceola) - FL Southern painted turtle (Chrysemys dorsalis) – AR Western painted turtle (Chrysemys picta) – IA, MO, OH, OK Spotted turtle (Clemmys gutatta) - FL, GA, OH Florida chicken turtle (Deirochelys reticularia chrysea) – FL Western chicken turtle (Deirochelys reticularia miaria) – AR, FL, GA, KY, MO, OK, TN, TX Barbour’s map turtle (Graptemys barbouri) - FL, GA Cagle’s map turtle (Graptemys caglei) - TX Escambia map turtle (Graptemys ernsti) – FL Common map turtle (Graptemys geographica) – AR, GA, OH, OK Ouachita map turtle (Graptemys ouachitensis) – AR, GA, OH, OK, TX Sabine map turtle (Graptemys ouachitensis sabinensis) – TX False map turtle (Graptemys pseudogeographica) – MO, OK, TX Mississippi map turtle (Graptemys pseuogeographica kohnii) – AR, TX Alabama map turtle (Graptemys pulchra) – GA Texas map turtle (Graptemys versa) - TX Striped mud turtle (Kinosternon baurii) – FL, GA, SC Yellow mud turtle (Kinosternon flavescens) – OK, TX Common mud turtle (Kinosternon subrubrum) – AR, FL, GA, OK, TX Alligator snapping turtle (Macrochelys temminckii) – AR, FL, GA, LA, MO, TX Diamond-back terrapin (Malaclemys terrapin) – FL, GA, LA, SC, TX River cooter (Pseudemys concinna) – AR, FL, -

Invasion of the Turtles? Wageningen Approach
Alterra is part of the international expertise organisation Wageningen UR (University & Research centre). Our mission is ‘To explore the potential of nature to improve the quality of life’. Within Wageningen UR, nine research institutes – both specialised and applied – have joined forces with Wageningen University and Van Hall Larenstein University of Applied Sciences to help answer the most important questions in the domain of healthy food and living environment. With approximately 40 locations (in the Netherlands, Brazil and China), 6,500 members of staff and 10,000 students, Wageningen UR is one of the leading organisations in its domain worldwide. The integral approach to problems and the cooperation between the exact sciences and the technological and social disciplines are at the heart of the Invasion of the turtles? Wageningen Approach. Alterra is the research institute for our green living environment. We offer a combination of practical and scientific Exotic turtles in the Netherlands: a risk assessment research in a multitude of disciplines related to the green world around us and the sustainable use of our living environment, such as flora and fauna, soil, water, the environment, geo-information and remote sensing, landscape and spatial planning, man and society. Alterra report 2186 ISSN 1566-7197 More information: www.alterra.wur.nl/uk R.J.F. Bugter, F.G.W.A. Ottburg, I. Roessink, H.A.H. Jansman, E.A. van der Grift and A.J. Griffioen Invasion of the turtles? Commissioned by the Invasive Alien Species Team of the Food and Consumer Product Safety Authority Invasion of the turtles? Exotic turtles in the Netherlands: a risk assessment R.J.F. -

Comparing Interrelated Stress Measures to Differentiate Acute and Chronic Stress in Urban Freshwater Turtles
Rollins College Rollins Scholarship Online Honors Program Theses Spring 2021 Comparing interrelated stress measures to differentiate acute and chronic stress in urban freshwater turtles Katherine Caldwell [email protected] Follow this and additional works at: https://scholarship.rollins.edu/honors Part of the Comparative and Evolutionary Physiology Commons, Fresh Water Studies Commons, and the Zoology Commons Recommended Citation Caldwell, Katherine, "Comparing interrelated stress measures to differentiate acute and chronic stress in urban freshwater turtles" (2021). Honors Program Theses. 148. https://scholarship.rollins.edu/honors/148 This Open Access is brought to you for free and open access by Rollins Scholarship Online. It has been accepted for inclusion in Honors Program Theses by an authorized administrator of Rollins Scholarship Online. For more information, please contact [email protected]. Comparing interrelated stress measures to differentiate acute and chronic stress in urban freshwater turtles Katie Caldwell Honors Thesis 2021 2 Abstract As urban areas grow in size and number, there are increasing impacts on the wildlife. While some species adapt, others cannot thrive in these conditions. Stress is the biological response to a stimulus that threatens homeostasis, which can be used to understand the condition of an individual. While acute stress is beneficial for survival, chronic stress can lead to deleterious impacts. A majority of turtles can be classified as declining in number or endangered, yet there is little understanding of their physiological stress response. This study examined how various stress measures—including corticosterone (CORT) in plasma, nails and leeches, metabolic measures, H:L ratio, and parasite prevalence—are related. We sampled freshwater turtles from two locations in the city of Winter Park, Florida, over a period of seven months. -

The Natural History & Distribution of Riverine Turtles in West Virginia
Marshall University Marshall Digital Scholar Theses, Dissertations and Capstones 2010 The aN tural History & Distribution of Riverine Turtles in West Virginia Linh Diem Phu Follow this and additional works at: http://mds.marshall.edu/etd Part of the Aquaculture and Fisheries Commons, and the Terrestrial and Aquatic Ecology Commons Recommended Citation Phu, Linh Diem, "The aN tural History & Distribution of Riverine Turtles in West Virginia" (2010). Theses, Dissertations and Capstones. Paper 787. This Thesis is brought to you for free and open access by Marshall Digital Scholar. It has been accepted for inclusion in Theses, Dissertations and Capstones by an authorized administrator of Marshall Digital Scholar. For more information, please contact [email protected]. The Natural History & Distribution of Riverine Turtles in West Virginia Thesis submitted to the Graduate College of Marshall University In partial fulfillment of the requirements for the degree of Master of Science in Biological Sciences By Linh Diem Phu Dr. Thomas K. Pauley, Ph.D., Committee Chairperson Dr. Dan Evans, Ph.D. Dr. Suzanne Strait, Ph.D. Marshall University May 2010 Abstract Turtles are unique evolutionary marvels that evolved from amphibians and developed their protective shelled form more than 200 million years ago. In West Virginia, there are 10 native species of turtles, 9 of which are aquatic. Most of these aquatic turtles feed on carrion and dead plant matter, in the water and essentially "clean" our water systems. Turtles are long-lived animals with sensitive life stages that can serve as both long-term and short-term bioindicators of environmental health. With the increase in commercial trade, habitat fragmentation, degradation, destruction, there has been a marked decline in turtle species. -

Invasion of the Turtles? Wageningen Approach
Alterra is part of the international expertise organisation Wageningen UR (University & Research centre). Our mission is ‘To explore the potential of nature to improve the quality of life’. Within Wageningen UR, nine research institutes – both specialised and applied – have joined forces with Wageningen University and Van Hall Larenstein University of Applied Sciences to help answer the most important questions in the domain of healthy food and living environment. With approximately 40 locations (in the Netherlands, Brazil and China), 6,500 members of staff and 10,000 students, Wageningen UR is one of the leading organisations in its domain worldwide. The integral approach to problems and the cooperation between the exact sciences and the technological and social disciplines are at the heart of the Invasion of the turtles? Wageningen Approach. Alterra is the research institute for our green living environment. We offer a combination of practical and scientific Exotic turtles in the Netherlands: a risk assessment research in a multitude of disciplines related to the green world around us and the sustainable use of our living environment, such as flora and fauna, soil, water, the environment, geo-information and remote sensing, landscape and spatial planning, man and society. Alterra report 2186 ISSN 1566-7197 More information: www.alterra.wur.nl/uk R.J.F. Bugter, F.G.W.A. Ottburg, I. Roessink, H.A.H. Jansman, E.A. van der Grift and A.J. Griffioen Invasion of the turtles? Commissioned by the Invasive Alien Species Team of the Food and Consumer Product Safety Authority Invasion of the turtles? Exotic turtles in the Netherlands: a risk assessment R.J.F. -

Proposed Amendment to 21CFR124021
Richard Fife 8195 S. Valley Vista Drive Hereford, AZ 85615 December 07, 2015 Division of Dockets Management Food and Drug Administration 5630 Fishers Lane, rm. 1061 Rockville, MD 20852 Reference: Docket Number FDA-2013-S-0610 Proposed Amendment to Code of Federal Regulations Title 21, Volume 8 Revised as of April 1, 2015 21CFR Sec.1240.62 Dear Dr. Stephen Ostroff, M.D., Acting Commissioner: Per discussion with the Division of Dockets Management staff on November 10, 2015 Environmental and Economic impact statements are not required for petitions submitted under 21CFR Sec.1240.62 CITIZEN PETITION December 07, 2015 ACTION REQUESTED: I propose an amendment to 21CFR Sec.1240.62 (see exhibit 1) as allowed by Section (d) Petitions as follows: Amend section (c) Exceptions. The provisions of this section are not applicable to: By adding the following two (2) exceptions: (5) The sale, holding for sale, and distribution of live turtles and viable turtle eggs, which are sold for a retail value of $75 or more (not to include any additional turtle related apparatuses, supplies, cages, food, or other turtle related paraphernalia). This dollar amount should be reviewed every 5 years or more often, as deemed necessary by the department in order to make adjustments for inflation using the US Department of Labor, Bureau of labor Statistics, Consumer Price Index. (6) The sale, holding for sale, and distribution of live turtles and viable turtle eggs, which are listed by the International Union for Conservation of Nature and Natural Resources (IUCN) Red List as Extinct In Wild, Critically Endangered, Endangered, or Vulnerable (IUCN threatened categorizes). -
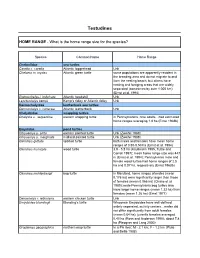
Home Range Testudines
Testudines HOME RANGE - What is the home range size for the species? Species Common Name Home Range Cheloniidae sea turtles Caretta c. caretta Atlantic loggerhead Unk Chelonia m. mydas Atlantic green turtle some populations are apparently resident in the breeding area and do not migrate to and from the nesting beach, but others have nesting and foraging areas that are widely separated (sometimes by over 1,000 km) (Ernst et al. 1994) Eretmochelys i. imbricata Atlantic hawksbill Unk Lepidochelys kempii Kemp’s ridley or Atlantic ridley Unk Dermochelyidae leatherback sea turtles Dermochelys c. coriacea Atlantic leatherback Unk Chelydridae snapping turtles Chelydra s. serpentina eastern snapping turtle in Pennsylvanina, nine adults…had estimated home ranges averaging 1.8 ha (Ernst 1968b) Emydidae pond turtles Chrysemys p. picta eastern painted turtle Unk (Zweifel 1989) Chrysemys p. marginata midland painted turtle Unk (Zweifel 1989) Clemmys guttata spotted turtle both males and females have mean home ranges of 0.50-0.53 ha (Ernst et al. 1994) Clemmys insculpta wood turtle 3.9 - 5.8 ha (Kaufmann 1995, Tuttle and Carroll 1997); mean home range size was 447 m (Ernst et al. 1994); Pennsylvania male and female wood turtles had home ranges of 2.5 ha and 0.07 ha, respectively (Ernst 1968b) Clemmys muhlenbergii bog turtle in Maryland, home ranges of males (mean 0.176 ha) were significantly larger than those of females (mean 0.066 ha) (Chase et al. 1989); male Pennsylvania bog turtles also have larger home ranges (mean 1.33 ha) than females (mean 1.26 ha) (Ernst 1977) Deirochelys r. -

Alien Invasive Species Profile
NOBANIS –Invasive Alien Species Fact Sheet Trachemys scripta Author of this fact sheet: Henrik Bringsøe, Nordisk Herpetologisk Forening (Nordic Herpetological Society), Irisvej 8, DK-4600 Køge, Denmark; +45 5666 3023; E-mail: [email protected] Bibliographical reference – how to cite this fact sheet: Bringsøe, H. (2006): NOBANIS – Invasive Alien Species Fact Sheet – Trachemys scripta. – From: Online Database of the North European and Baltic Network on Invasive Alien Species – NOBANIS www.nobanis.org, Date of access x/x/200x. Species description Scientific name: Trachemys scripta (Schoepff, 1792), Testudines, Emydidae Synonyms: Chrysemys scripta: Boulenger 1889 Pseudemys scripta: Jordan 1899 Common names: Slider (US), Nordamerikansk terrapin (DK), Buchstaben-Schmuckschildkröte (DE), punakorvakilpikonna (FI), Krasnoukhaya cherepakha (красноухая черепаха) (RU), żółw ozdobny (PL), sarkanausu bruņurupucis (LV) , punakõrv-ilukilpkonn (EE). Subspecies and their common names: Trachemys scripta scripta (Schoepff, 1792): yellow-bellied slider (US), yellow-bellied terrapin (GB), guløretguløret terrapin or gulbuget terrapin (DK), gulbukad vattensköldpadda (SE), eigentliche Buchstaben-Schmuckschildkröte or Gelbbauch-Schmuckschildkröte (DE), żółw żółtolicy or żółw żółtouchy or żółw żółtobrzuchy (PL). Trachemys scripta elegans (Wied, 1839): red-eared slider (US), red-eared terrapin (GB), rødøret terrapin (DK), rödörad vattensköldpadda (SE), rødøreterrapin (NO), punakorvakilpikonna (FI), Rotwangen-Schmuckschildkröte (DE), żółw czerwonolicy or żółw czerwonouchy (PL), punakõrv-ilukilpkonn (EE), raudonausis vėžlys or raudonskruostis vėžlys (LT). Trachemys scripta troostii (Holbrook, 1836): Cumberland slider (US), Troost’s terrapin (GB), cumberland-terrapin (DK), Gelbwangen-Schmuckschildkröte or Troosts Schmuckschildkröte (DE). Fig 1. Adult female Trachemys scripta elegans basking on a rock in the Botanical Garden of Copenhagen, Denmark. The conspicuous reddish orange to red stripe behind the eye is characteristic of that subspecies, photo by Henrik Bringsøe. -

Red-Eared Slider
INVASIVE SPECIES ALERT! RED-EARED SLIDER (Trachemys scripta elegans) REPORT INVASIVE SPECIES NATIVE RANGE www.reportinvasives.ca The Red-Eared Slider is native to southeastern United States and south through Mexico, Central America and Brazil. DESCRIPTION Photo: Kristiina Ovaska Red-Eared Sliders… Are green with fine yellow stripes on their head, neck, limbs, and tail Have distinguishing red ‘ear’ patches (may fade to dark brown or yellow) on sides of the head Have a top shell (carapace) that is dome shaped and that is dark green with light and dark markings Have a carapace length up to 28 cm (11”) Are yellow on bottom of shell (plastron) with dark blotched markings For more information refer to the BC Turtle Watch ID Guide http://hat.bc.ca/attachments/Turtle-IDGuide-May2008.pdf PRIMARY IMPACT: Red-Eared Sliders are WHY SHOULD WE CARE? known carriers of Red-Eared Sliders… diseases that are harmful May compete with native turtles for food and habitat May carry diseases contracted from other turtles in the pet to humans and wildlife. trade that they can transfer to native turtles * In the state of Washington, populations of the native turtle Actinemys marmorata were seriously impacted by a respiratory infection in 1990 and T. scripta was the likely vector (Hays et al. 1999) Pet Red-eared Sliders have been known carriers of Salmonella that is harmful to humans DID YOU KNOW? Sliders have a fascinating kind of feeding BIOLOGY & SPREAD system called neustophagia, which is Due to release by pet owners, Red-Eared Sliders have spread into similar to the feeding style of the baleen ponds and wetlands of B.C.