DNA Diagnostic Lab at JOHNS HOPKINS Test Requisition Part I of II
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Next Generation Sequencing Panels for Disorders of Sex Development
Next Generation Sequencing Panels for Disorders of Sex Development Disorders of Sex Development – Overview Disorders of sex development (DSDs) occur when sex development does not follow the course of typical male or female patterning. Types of DSDs include congenital development of ambiguous genitalia, disjunction between the internal and external sex anatomy, incomplete development of the sex anatomy, and abnormalities of the development of gonads (such as ovotestes or streak ovaries) (1). Sex chromosome anomalies including Turner syndrome and Klinefelter syndrome as well as sex chromosome mosaicism are also considered to be DSDs. DSDs can be caused by a wide range of genetic abnormalities (2). Determining the etiology of a patient’s DSD can assist in deciding gender assignment, provide recurrence risk information for future pregnancies, and can identify potential health problems such as adrenal crisis or gonadoblastoma (1, 3). Sex chromosome aneuploidy and copy number variation are common genetic causes of DSDs. For this reason, chromosome analysis and/or microarray analysis typically should be the first genetic analysis in the case of a patient with ambiguous genitalia or other suspected disorder of sex development. Identifying whether a patient has a 46,XY or 46,XX karyotype can also be helpful in determining appropriate additional genetic testing. Abnormal/Ambiguous Genitalia Panel Our Abnormal/Ambiguous Genitalia Panel includes mutation analysis of 72 genes associated with both syndromic and non-syndromic DSDs. This comprehensive panel evaluates a broad range of genetic causes of ambiguous or abnormal genitalia, including conditions in which abnormal genitalia are the primary physical finding as well as syndromic conditions that involve abnormal genitalia in addition to other congenital anomalies. -

The Genetic Basis for Skeletal Diseases
insight review articles The genetic basis for skeletal diseases Elazar Zelzer & Bjorn R. Olsen Harvard Medical School, Department of Cell Biology, 240 Longwood Avenue, Boston, Massachusetts 02115, USA (e-mail: [email protected]) We walk, run, work and play, paying little attention to our bones, their joints and their muscle connections, because the system works. Evolution has refined robust genetic mechanisms for skeletal development and growth that are able to direct the formation of a complex, yet wonderfully adaptable organ system. How is it done? Recent studies of rare genetic diseases have identified many of the critical transcription factors and signalling pathways specifying the normal development of bones, confirming the wisdom of William Harvey when he said: “nature is nowhere accustomed more openly to display her secret mysteries than in cases where she shows traces of her workings apart from the beaten path”. enetic studies of diseases that affect skeletal differentiation to cartilage cells (chondrocytes) or bone cells development and growth are providing (osteoblasts) within the condensations. Subsequent growth invaluable insights into the roles not only of during the organogenesis phase generates cartilage models individual genes, but also of entire (anlagen) of future bones (as in limb bones) or membranous developmental pathways. Different mutations bones (as in the cranial vault) (Fig. 1). The cartilage anlagen Gin the same gene may result in a range of abnormalities, are replaced by bone and marrow in a process called endo- and disease ‘families’ are frequently caused by mutations in chondral ossification. Finally, a process of growth and components of the same pathway. -

Mutations Within Or Upstream of the Basic Helixð Loopð Helix Domain of the TWIST Gene Are Specific to Saethre-Chotzen Syndrome
European Journal of Human Genetics (1999) 7, 27–33 © 1999 Stockton Press All rights reserved 1018–4813/99 $12.00 t http://www.stockton-press.co.uk/ejhg ARTICLES Mutations within or upstream of the basic helix–loop–helix domain of the TWIST gene are specific to Saethre-Chotzen syndrome Vincent El Ghouzzi, Elisabeth Lajeunie, Martine Le Merrer, Val´erie Cormier-Daire, Dominique Renier, Arnold Munnich and Jacky Bonaventure Unit´e de Recherches sur les Handicaps G´en´etiques de l’Enfant, Institut Necker, Paris, France Saethre-Chotzen syndrome (ACS III) is an autosomal dominant craniosynostosis syndrome recently ascribed to mutations in the TWIST gene, a basic helix–loop–helix (b-HLH) transcription factor regulating head mesenchyme cell development during cranial neural tube formation in mouse. Studying a series of 22 unrelated ACS III patients, we have found TWIST mutations in 16/22 cases. Interestingly, these mutations consistently involved the b-HLH domain of the protein. Indeed, mutant genotypes included frameshift deletions/insertions, nonsense and missense mutations, either truncating or disrupting the b-HLH motif of the protein. This observation gives additional support to the view that most ACS III cases result from loss-of-function mutations at the TWIST locus. The P250R recurrent FGFR 3 mutation was found in 2/22 cases presenting mild clinical manifestations of the disease but 4/22 cases failed to harbour TWIST or FGFR 3 mutations. Clinical re-examination of patients carrying TWIST mutations failed to reveal correlations between the mutant genotype and severity of the phenotype. Finally, since no TWIST mutations were detected in 40 cases of isolated coronal craniosynostosis, the present study suggests that TWIST mutations are specific to Saethre- Chotzen syndrome. -

MECHANISMS in ENDOCRINOLOGY: Novel Genetic Causes of Short Stature
J M Wit and others Genetics of short stature 174:4 R145–R173 Review MECHANISMS IN ENDOCRINOLOGY Novel genetic causes of short stature 1 1 2 2 Jan M Wit , Wilma Oostdijk , Monique Losekoot , Hermine A van Duyvenvoorde , Correspondence Claudia A L Ruivenkamp2 and Sarina G Kant2 should be addressed to J M Wit Departments of 1Paediatrics and 2Clinical Genetics, Leiden University Medical Center, PO Box 9600, 2300 RC Leiden, Email The Netherlands [email protected] Abstract The fast technological development, particularly single nucleotide polymorphism array, array-comparative genomic hybridization, and whole exome sequencing, has led to the discovery of many novel genetic causes of growth failure. In this review we discuss a selection of these, according to a diagnostic classification centred on the epiphyseal growth plate. We successively discuss disorders in hormone signalling, paracrine factors, matrix molecules, intracellular pathways, and fundamental cellular processes, followed by chromosomal aberrations including copy number variants (CNVs) and imprinting disorders associated with short stature. Many novel causes of GH deficiency (GHD) as part of combined pituitary hormone deficiency have been uncovered. The most frequent genetic causes of isolated GHD are GH1 and GHRHR defects, but several novel causes have recently been found, such as GHSR, RNPC3, and IFT172 mutations. Besides well-defined causes of GH insensitivity (GHR, STAT5B, IGFALS, IGF1 defects), disorders of NFkB signalling, STAT3 and IGF2 have recently been discovered. Heterozygous IGF1R defects are a relatively frequent cause of prenatal and postnatal growth retardation. TRHA mutations cause a syndromic form of short stature with elevated T3/T4 ratio. Disorders of signalling of various paracrine factors (FGFs, BMPs, WNTs, PTHrP/IHH, and CNP/NPR2) or genetic defects affecting cartilage extracellular matrix usually cause disproportionate short stature. -

Blueprint Genetics Comprehensive Growth Disorders / Skeletal
Comprehensive Growth Disorders / Skeletal Dysplasias and Disorders Panel Test code: MA4301 Is a 374 gene panel that includes assessment of non-coding variants. This panel covers the majority of the genes listed in the Nosology 2015 (PMID: 26394607) and all genes in our Malformation category that cause growth retardation, short stature or skeletal dysplasia and is therefore a powerful diagnostic tool. It is ideal for patients suspected to have a syndromic or an isolated growth disorder or a skeletal dysplasia. About Comprehensive Growth Disorders / Skeletal Dysplasias and Disorders This panel covers a broad spectrum of diseases associated with growth retardation, short stature or skeletal dysplasia. Many of these conditions have overlapping features which can make clinical diagnosis a challenge. Genetic diagnostics is therefore the most efficient way to subtype the diseases and enable individualized treatment and management decisions. Moreover, detection of causative mutations establishes the mode of inheritance in the family which is essential for informed genetic counseling. For additional information regarding the conditions tested on this panel, please refer to the National Organization for Rare Disorders and / or GeneReviews. Availability 4 weeks Gene Set Description Genes in the Comprehensive Growth Disorders / Skeletal Dysplasias and Disorders Panel and their clinical significance Gene Associated phenotypes Inheritance ClinVar HGMD ACAN# Spondyloepimetaphyseal dysplasia, aggrecan type, AD/AR 20 56 Spondyloepiphyseal dysplasia, Kimberley -

Blueprint Genetics Comprehensive Skeletal Dysplasias and Disorders
Comprehensive Skeletal Dysplasias and Disorders Panel Test code: MA3301 Is a 251 gene panel that includes assessment of non-coding variants. Is ideal for patients with a clinical suspicion of disorders involving the skeletal system. About Comprehensive Skeletal Dysplasias and Disorders This panel covers a broad spectrum of skeletal disorders including common and rare skeletal dysplasias (eg. achondroplasia, COL2A1 related dysplasias, diastrophic dysplasia, various types of spondylo-metaphyseal dysplasias), various ciliopathies with skeletal involvement (eg. short rib-polydactylies, asphyxiating thoracic dysplasia dysplasias and Ellis-van Creveld syndrome), various subtypes of osteogenesis imperfecta, campomelic dysplasia, slender bone dysplasias, dysplasias with multiple joint dislocations, chondrodysplasia punctata group of disorders, neonatal osteosclerotic dysplasias, osteopetrosis and related disorders, abnormal mineralization group of disorders (eg hypopohosphatasia), osteolysis group of disorders, disorders with disorganized development of skeletal components, overgrowth syndromes with skeletal involvement, craniosynostosis syndromes, dysostoses with predominant craniofacial involvement, dysostoses with predominant vertebral involvement, patellar dysostoses, brachydactylies, some disorders with limb hypoplasia-reduction defects, ectrodactyly with and without other manifestations, polydactyly-syndactyly-triphalangism group of disorders, and disorders with defects in joint formation and synostoses. Availability 4 weeks Gene Set Description -

Essential Genetics 5
Essential genetics 5 Disease map on chromosomes 例 Gaucher disease 単一遺伝子病 天使病院 Prader-Willi syndrome 隣接遺伝子症候群,欠失が主因となる疾患 臨床遺伝診療室 外木秀文 Trisomy 13 複数の遺伝子の重複によって起こる疾患 挿画 Koromo 遺伝子の座位あるいは欠失等の範囲を示す Copyright (c) 2010 Social Medical Corporation BOKOI All Rights Reserved. Disease map on chromosome 1 Gaucher disease Chromosome 1q21.1 1p36 deletion syndrome deletion syndrome Adrenoleukodystrophy, neonatal Cardiomyopathy, dilated, 1A Zellweger syndrome Charcot-Marie-Tooth disease Emery-Dreifuss muscular Hypercholesterolemia, familial dystrophy Hutchinson-Gilford progeria Ehlers-Danlos syndrome, type VI Muscular dystrophy, limb-girdle type Congenital disorder of Insensitivity to pain, congenital, glycosylation, type Ic with anhidrosis Diamond-Blackfan anemia 6 Charcot-Marie-Tooth disease Dejerine-Sottas syndrome Marshall syndrome Stickler syndrome, type II Chronic granulomatous disease due to deficiency of NCF-2 Alagille syndrome 2 Copyright (c) 2010 Social Medical Corporation BOKOI All Rights Reserved. Disease map on chromosome 2 Epiphyseal dysplasia, multiple Spondyloepimetaphyseal dysplasia Brachydactyly, type D-E, Noonan syndrome Brachydactyly-syndactyly syndrome Peters anomaly Synpolydactyly, type II and V Parkinson disease, familial Leigh syndrome Seizures, benign familial Multiple pterygium syndrome neonatal-infantile Escobar syndrome Ehlers-Danlos syndrome, Brachydactyly, type A1 type I, III, IV Waardenburg syndrome Rhizomelic chondrodysplasia punctata, type 3 Alport syndrome, autosomal recessive Split-hand/foot malformation Crigler-Najjar -

REVIEW ARTICLE Genetic Disorders of the Skeleton: a Developmental Approach
Am. J. Hum. Genet. 73:447–474, 2003 REVIEW ARTICLE Genetic Disorders of the Skeleton: A Developmental Approach Uwe Kornak and Stefan Mundlos Institute for Medical Genetics, Charite´ University Hospital, Campus Virchow, Berlin Although disorders of the skeleton are individually rare, they are of clinical relevance because of their overall frequency. Many attempts have been made in the past to identify disease groups in order to facilitate diagnosis and to draw conclusions about possible underlying pathomechanisms. Traditionally, skeletal disorders have been subdivided into dysostoses, defined as malformations of individual bones or groups of bones, and osteochondro- dysplasias, defined as developmental disorders of chondro-osseous tissue. In light of the recent advances in molecular genetics, however, many phenotypically similar skeletal diseases comprising the classical categories turned out not to be based on defects in common genes or physiological pathways. In this article, we present a classification based on a combination of molecular pathology and embryology, taking into account the importance of development for the understanding of bone diseases. Introduction grouping of conditions that have a common molecular origin but that have little in common clinically. For ex- Genetic disorders affecting the skeleton comprise a large ample, mutations in COL2A1 can result in such diverse group of clinically distinct and genetically heterogeneous conditions as lethal achondrogenesis type II and Stickler conditions. Clinical manifestations range from neonatal dysplasia, which is characterized by moderate growth lethality to only mild growth retardation. Although they retardation, arthropathy, and eye disease. It is now be- are individually rare, disorders of the skeleton are of coming increasingly clear that several distinct classifi- clinical relevance because of their overall frequency. -

Disorders of Sex Development Panel
Abnormal Genitalia/ Disorders of Sex Development Panel Test code: EN0201 Is a 62 gene panel that includes assessment of non-coding variants. Is ideal for patients presenting with ambiguous genitalia, patients suspected to have a disorder of sexual development and patients suspected to have congenital adrenal hyperplasia (CAH). About Abnormal Genitalia/ Disorders of Sex Development Disorders of sex development (DSD) are a group of congenital conditions characterized by problems in the course of gender patterning, gonadal and sex development. It has been estimated that 1% – 2% of live births have some aspect of DSD. Approximately 5% of infants with DSD have ambiguous genitalia and indeterminate sex at birth. However, the vast majority of these patients do not require corrective surgery. Patients with 46,XY DSD have often impaired androgen synthesis or action and may have normal female external genitalia, while patients with 46,XX DSD conditions have often androgen excess. In 46,XX females, congenital adrenal hyperplasia (CAH) caused by 21-hydroxylase deficiency (21-OHD) is the most common cause of DSD. The estimated prevalence of CAH is 1:10,000 and 90%-95% of cases are due to mutations in CYP21A2. The severity of the condition often depends on the residual enzyme activity subdiving CYP21A2 mutations in severe (classic phenotype, enzyme activity 0%-10%) and mild (non-classic, enzyme activity 20%-50%). Androgen insensitivity syndrome (AIS), caused by mutations in AR, is characterized by feminization of external genitalia and atypical sexual development in 46,XY individuals. The condition may be complete, partial or mild, depending on the level of androgen insensitivity. -
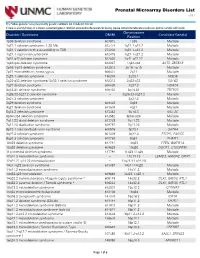
Prenatal Microarray Disorders List V19.1
Prenatal Microarray Disorders List v19.1 This "whole genome" array may identify genetic conditions not included in this list. If there is a family history of a known suspected genetic condition unrelated to the reason for testing, please contact the laboratory to discuss prior to sample submission. Chromosome Disorder / Syndrome OMIM Candidate Gene(s) Position 1p36 deletion syndrome 607872 1p36 Multiple 1q21.1 deletion syndrome, 1.35 Mb 612474 1q21.1-q21.2 Multiple 1q21.1 deletion with susceptibility to TAR 274000 1q21.1-q21.2 Multiple 1q21.1 duplication syndrome 612475 1q21.1-q21.2 Multiple 1q41-q42 deletion syndrome 612530 1q41-q42.12 Multiple 1q43-q44 deletion syndrome 612337 1q43-q44 AKT3, ZBTB18 2p16.1-p15 deletion syndrome 612513 2p16.1-p15 Multiple 2p21 microdeletion, homozygous 606407 2p21 Multiple 2q23.1 deletion syndrome 156200 2q23.1 MBD5 2q32-q33 deletion syndrome/ 2q33.1 deletion syndrome 612313 2q32-q33 SATB2 2q37 deletion syndrome 600430 2q37.3 HDAC4 3q13.31 deletion syndrome 615433 3q13.31 ZBTB20 3q26.33-3q27.2 deletion syndrome -- 3q26.33-3q27.2 Multiple 3q27.3 deletion syndrome -- 3q27.3 Multiple 3q29 deletion syndrome 609425 3q29 Multiple 4q21 deletion syndrome 613509 4q21 Multiple 5q14.3 deletion syndrome 613443 5q14.3 MEF2C 6pter-p24 deletion syndrome 612582 6pter-p24 Multiple 7q11.23 distal deletion syndrome 613729 7q11.23 Multiple 7q11.23 duplication syndrome 609757 7q11.23 Multiple 8p23.1 deletion/duplication syndrome 600576 8p23.1 GATA4 9q22.3 deletion syndrome 601309 9q22.3 PTCH1, FANCC 9q34.3 deletion syndrome -

Campomelic Dysplasia
Campomelic Dysplasia Overview Campomelic dysplasia is a rare form of bent-bone skeletal dysplasia that affects an estimated 1 in 40,000-200,000 people. It is complicated by respiratory issues and has therefore historically been considered a lethal disease, with most individuals not surviving past infancy. With proper medical care and guidance, campomelic dysplasia can be managed, although not without lasting orthopaedic and miscellaneous concerns. Campomelic dysplasia may be detecting using radiological techniques, physical exams and genetic testing. Clinical features include underdevelopment of the jaw, cleft palate, clubfoot, spine deformities and leg alignment issues. Treatment of symptoms may include monitoring and surgery by doctors who specialize in skeletal dysplasias. Heredity and genetics Campomelic dysplasia is caused by mutations in the SOX9 gene. This gene codes for a transcription factor — or protein — that attaches to genes to help them be expressed, particularly those involved in development of the skeleton and reproductive system. Under normal circumstances, there are two functioning copies of SOX9. Campomelic dysplasia occurs when one copy of the gene becomes defective — either through a point mutation on or near the gene, rearrangement of the chromosome or when the chromosome is deleted. Campomelic dysplasia tends to be less severe when the mutation involves a chromosomal abnormality instead of direct mutations of SOX9. Most cases of campomelic dysplasia happen from spontaneous mutations during fetal development. However, the disease can be inherited in an autosomal dominant fashion, meaning one copy of the defective gene is sufficient to cause the disease in the offspring. If an affected individual survives past reproductive age, they have a 50 percent chance of passing on the mutation to their child. -

Skeletal Dysplasia Panel Versie V1 (345 Genen) Centrum Voor Medische Genetica Gent
H9.1-OP2-B40: Genpanel Skeletal dysplasia, V1, in voege op 14/02/2020 Skeletal_dysplasia panel versie V1 (345 genen) Centrum voor Medische Genetica Gent Associated phenotype, OMIM phenotype ID, phenotype Gene OMIM gene ID mapping key and inheritance pattern Atrial fibrillation, familial, 12, 614050 (3), Autosomal dominant; ABCC9 601439 Cardiomyopathy, dilated, 1O, 608569 (3); Hypertrichotic osteochondrodysplasia, 239850 (3), Autosomal dominant Congenital heart defects and skeletal malformations syndrome, 617602 (3), Autosomal dominant; Leukemia, Philadelphia ABL1 189980 chromosome-positive, resistant to imatinib, 608232 (3), Somatic mutation Short stature and advanced bone age, with or without early-onset osteoarthritis and/or osteochondritis dissecans, 165800 (3), ACAN 155760 Autosomal dominant; Spondyloepimetaphyseal dysplasia, aggrecan type, 612813 (3), Autosomal recessive; ?Spondyloepiphyseal dysplasia, Kimberley type, 608361 (3), Autosomal dominant Spondyloenchondrodysplasia with immune dysregulation, 607944 ACP5 171640 (3), Autosomal recessive Fibrodysplasia ossificans progressiva, 135100 (3), Autosomal ACVR1 102576 dominant Weill-Marchesani syndrome 1, recessive, 277600 (3), Autosomal ADAMTS10 608990 recessive Weill-Marchesani 4 syndrome, recessive, 613195 (3), Autosomal ADAMTS17 607511 recessive ADAMTSL2 612277 Geleophysic dysplasia 1, 231050 (3), Autosomal recessive AFF4 604417 CHOPS syndrome, 616368 (3), Autosomal dominant AGA 613228 Aspartylglucosaminuria, 208400 (3), Autosomal recessive Rhizomelic chondrodysplasia punctata,