Some Bioinformatic Analyses of Human GDAP1 Gene Expression
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

An Order Estimation Based Approach to Identify Response Genes
AN ORDER ESTIMATION BASED APPROACH TO IDENTIFY RESPONSE GENES FOR MICRO ARRAY TIME COURSE DATA A Thesis Presented to The Faculty of Graduate Studies of The University of Guelph by ZHIHENG LU In partial fulfilment of requirements for the degree of Doctor of Philosophy September, 2008 © Zhiheng Lu, 2008 Library and Bibliotheque et 1*1 Archives Canada Archives Canada Published Heritage Direction du Branch Patrimoine de I'edition 395 Wellington Street 395, rue Wellington Ottawa ON K1A0N4 Ottawa ON K1A0N4 Canada Canada Your file Votre reference ISBN: 978-0-494-47605-5 Our file Notre reference ISBN: 978-0-494-47605-5 NOTICE: AVIS: The author has granted a non L'auteur a accorde une licence non exclusive exclusive license allowing Library permettant a la Bibliotheque et Archives and Archives Canada to reproduce, Canada de reproduire, publier, archiver, publish, archive, preserve, conserve, sauvegarder, conserver, transmettre au public communicate to the public by par telecommunication ou par Plntemet, prefer, telecommunication or on the Internet, distribuer et vendre des theses partout dans loan, distribute and sell theses le monde, a des fins commerciales ou autres, worldwide, for commercial or non sur support microforme, papier, electronique commercial purposes, in microform, et/ou autres formats. paper, electronic and/or any other formats. The author retains copyright L'auteur conserve la propriete du droit d'auteur ownership and moral rights in et des droits moraux qui protege cette these. this thesis. Neither the thesis Ni la these ni des extraits substantiels de nor substantial extracts from it celle-ci ne doivent etre imprimes ou autrement may be printed or otherwise reproduits sans son autorisation. -
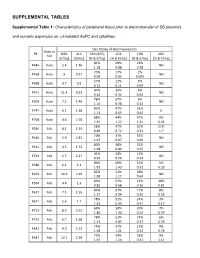
Supplemental Tables
SUPPLEMENTAL TABLES Supplemental Table 1: Characteristics of peripheral blood prior to electrotransfer of SB plasmids and numeric expansion on γ-irradiated AaPC and cytokines Day 0 (Day of electroporation) Auto or P# WBC ALC CD3 (ATC) CD4 CD8 ABC Allo [K/mL] [K/mL] [% & K/mL] [% & K/mL] [% & K/mL] [% & K/mL] 81% 60% 19% P446 Auto 5.4 1.46 ND 1.18 0.88 0.28 23% 17% 2% P458 Auto 9 0.27 ND 0.06 0.05 0.005 17% 12% 5% P468 Auto 4.7 0.9 ND 0.15 0.11 0.04 47% 35% 5% P471 Auto 11.4 0.93 ND 0.44 0.32 0.04 78% 67% 9% P509 Auto 7.2 1.46 ND 1.14 0.98 0.13 82% 47% 32% P747 Auto 6.1 1.38 0 1.13 0.65 0.44 88% 44% 47% 0% P708 Auto 4.6 2.78 2.45 1.22 1.31 0.05 58% 47% 20% 21% P396 Allo 8.1 1.54 0.89 0.72 0.31 1.7 70% 31% 32% P410 Allo 5.9 2.81 ND 1.97 0.87 0.90 80% 48% 32% P411 Allo 4.5 1.72 ND 1.38 0.83 0.55 41% 24% 15% P513 Allo 6.7 2.27 ND 0.93 0.54 0.34 86% 68% 15% 5% P580 Allo 6.1 2.1 1.81 1.43 0.32 0.29 82% 51% 28% P459 Allo 10.6 2.29 ND 1.88 1.17 0.64 64% 52% 12% 18% P564 Allo 4.4 1.3 0.83 0.68 0.16 0.81 82% 61% 17% 8% P617 Allo 7.1 1.55 1.27 0.94 0.26 0.58 78% 53% 24% 3% P671 Allo 5.4 1.7 1.33 0.90 0.41 0.17 69% 50% 20% 7% P723 Allo 8.6 2.61 1.80 1.30 0.52 0.59 78% 52% 22% 6% P732 Allo 6.7 1.68 1.31 0.87 0.37 0.39 74% 57% 15% 9% P641 Allo 9.0 2.13 1.58 1.21 0.32 0.78 73% 54% 18% 9% P647 Allo 12.1 2.29 1.67 1.24 0.41 1.11 75% 59% 11% P716 Allo 3.0 1.83 0 1.37 1.08 0.20 87% 73% 12% 1% P718 Allo 6.2 1.99 1.73 1.45 0.24 0.07 83% 69% 13% 6% P771 Allo 13.1 3.23 2.68 2.23 0.42 0.79 73% 42% 27% 6% P783 Allo 8.1 1.54 1.12 0.65 0.42 0.52 WBC = white blood -

P53 — 30 Yreviewsears On
FOCUS ON P53 — 30 YREVIEWSEARS ON p53 — a Jack of all trades but master of none Melissa R. Junttila and Gerard I. Evan Abstract | Cancers are rare because their evolution is actively restrained by a range of tumour suppressors. Of these p53 seems unusually crucial as either it or its attendant upstream or downstream pathways are inactivated in virtually all cancers. p53 is an evolutionarily ancient coordinator of metazoan stress responses. Its role in tumour suppression is likely to be a relatively recent adaptation, which is only necessary when large, long-lived organisms acquired the sufficient size and somatic regenerative capacity to necessitate specific mechanisms to reign in rogue proliferating cells. However, such evolutionary reappropriation of this venerable transcription factor entails compromises that restrict its efficacy as a tumour suppressor. Cnidarians–bilaterians Cancer is a genetic pathology that arises in the adult tissues questions are important for several reasons. Knowing Cinidarians comprise an animal of long-lived organisms, such as vertebrates, whose tis- how p53 discriminates between normal and tumour cells phylum of ~9,000 radially sues retain a substantial regenerative capacity throughout might point to attributes of cancer cells that qualitatively symmetrical, mostly marine life and, consequently, whose somatic cells accumulate distinguish them from normal somatic cells and could organisms. Most other animals are bilaterally symmetrical mutations. Occasionally, such mutations corrupt the be used as tumour-specific targets. Knowing what sig- and are classed as bilateria. regulatory mechanisms that suppress untoward somatic nals drive selection for loss of p53 function in cancers The cnidarians and bilaterians cell proliferation, survival and migration, resulting in the would help us understand what constrains and dictates last shared a common ancestor progressive outgrowth of a somatic clone. -

Mitoxplorer, a Visual Data Mining Platform To
mitoXplorer, a visual data mining platform to systematically analyze and visualize mitochondrial expression dynamics and mutations Annie Yim, Prasanna Koti, Adrien Bonnard, Fabio Marchiano, Milena Dürrbaum, Cecilia Garcia-Perez, José Villaveces, Salma Gamal, Giovanni Cardone, Fabiana Perocchi, et al. To cite this version: Annie Yim, Prasanna Koti, Adrien Bonnard, Fabio Marchiano, Milena Dürrbaum, et al.. mitoXplorer, a visual data mining platform to systematically analyze and visualize mitochondrial expression dy- namics and mutations. Nucleic Acids Research, Oxford University Press, 2020, 10.1093/nar/gkz1128. hal-02394433 HAL Id: hal-02394433 https://hal-amu.archives-ouvertes.fr/hal-02394433 Submitted on 4 Dec 2019 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Distributed under a Creative Commons Attribution| 4.0 International License Nucleic Acids Research, 2019 1 doi: 10.1093/nar/gkz1128 Downloaded from https://academic.oup.com/nar/advance-article-abstract/doi/10.1093/nar/gkz1128/5651332 by Bibliothèque de l'université la Méditerranée user on 04 December 2019 mitoXplorer, a visual data mining platform to systematically analyze and visualize mitochondrial expression dynamics and mutations Annie Yim1,†, Prasanna Koti1,†, Adrien Bonnard2, Fabio Marchiano3, Milena Durrbaum¨ 1, Cecilia Garcia-Perez4, Jose Villaveces1, Salma Gamal1, Giovanni Cardone1, Fabiana Perocchi4, Zuzana Storchova1,5 and Bianca H. -

Integration of 198 Chip-Seq Datasets Reveals Human Cis-Regulatory Regions
Integration of 198 ChIP-seq datasets reveals human cis-regulatory regions Hamid Bolouri & Larry Ruzzo Thanks to Stephen Tapscott, Steven Henikoff & Zizhen Yao Slides will be available from: http://labs.fhcrc.org/bolouri/ Email [email protected] for manuscript (Bolouri & Ruzzo, submitted) Kleinjan & van Heyningen, Am. J. Hum. Genet., 2005, (76)8–32 Epstein, Briefings in Func. Genom. & Protemoics, 2009, 8(4)310-16 Regulation of SPi1 (Sfpi1, PU.1 protein) expression – part 1 miR155*, miR569# ~750nt promoter ~250nt promoter The antisense RNA • causes translational stalling • has its own promoter • requires distal SPI1 enhancer • is transcribed with/without SPI1. # Hikami et al, Arthritis & Rheumatism, 2011, 63(3):755–763 * Vigorito et al, 2007, Immunity 27, 847–859 Ebralidze et al, Genes & Development, 2008, 22: 2085-2092. Regulation of SPi1 expression – part 2 (mouse coordinates) Bidirectional ncRNA transcription proportional to PU.1 expression PU.1/ELF1/FLI1/GLI1 GATA1 GATA1 Sox4/TCF/LEF PU.1 RUNX1 SP1 RUNX1 RUNX1 SP1 ELF1 NF-kB SATB1 IKAROS PU.1 cJun/CEBP OCT1 cJun/CEBP 500b 500b 500b 500b 500b 750b 500b -18Kb -14Kb -12Kb -10Kb -9Kb Chou et al, Blood, 2009, 114: 983-994 Hoogenkamp et al, Molecular & Cellular Biology, 2007, 27(21):7425-7438 Zarnegar & Rothenberg, 2010, Mol. & cell Biol. 4922-4939 An NF-kB binding-site variant in the SPI1 URE reduces PU.1 expression & is GGGCCTCCCC correlated with AML GGGTCTTCCC Bonadies et al, Oncogene, 2009, 29(7):1062-72. SATB1 binding site A distal single nucleotide polymorphism alters long- range regulation of the PU.1 gene in acute myeloid leukemia Steidl et al, J Clin Invest. -

BMC Genomics Biomed Central
BMC Genomics BioMed Central Methodology article Open Access "NeuroStem Chip": a novel highly specialized tool to study neural differentiation pathways in human stem cells Sergey V Anisimov*, Nicolaj S Christophersen, Ana S Correia, Jia-Yi Li and Patrik Brundin Address: Neuronal Survival Unit, Wallenberg Neuroscience Center, Lund University, 221 84 Lund, Sweden Email: Sergey V Anisimov* - [email protected]; Nicolaj S Christophersen - [email protected]; Ana S Correia - [email protected]; Jia-Yi Li - [email protected]; Patrik Brundin - [email protected] * Corresponding author Published: 8 February 2007 Received: 21 September 2006 Accepted: 8 February 2007 BMC Genomics 2007, 8:46 doi:10.1186/1471-2164-8-46 This article is available from: http://www.biomedcentral.com/1471-2164/8/46 © 2007 Anisimov et al; licensee BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Abstract Background: Human stem cells are viewed as a possible source of neurons for a cell-based therapy of neurodegenerative disorders, such as Parkinson's disease. Several protocols that generate different types of neurons from human stem cells (hSCs) have been developed. Nevertheless, the cellular mechanisms that underlie the development of neurons in vitro as they are subjected to the specific differentiation protocols are often poorly understood. Results: We have designed a focused DNA (oligonucleotide-based) large-scale microarray platform (named "NeuroStem Chip") and used it to study gene expression patterns in hSCs as they differentiate into neurons. -

Prostate News
Prostate Cancer and Prostatic Diseases (2005) 8, 2–6 & 2005 Nature Publishing Group All rights reserved 1365-7852/05 $30.00 www.nature.com/pcan Research Highlights Prostate News Prostate Cancer and Prostatic Diseases (2005) 8, 2–6. doi:10.1038/sj.pcan.4500793 Pokemon’s central role in cancer progression of prostate cancer. Epigenetic changes are reversible, which make them attractive targets for cancer development treatment with modulators that demethylate DNA and Researchers at New York’s Memorial Sloan-Kettering inhibit histone deacetylases, leading to reactivation of Cancer Center report novel findings in Nature regarding silenced genes. DNA methylation and histone modifica- a new cellular oncogene implicated in the development tions are important epigenetic mechanisms of gene of human cancers, including prostate neoplasia. Poke- regulation, and both play essential roles in tumor mon, a pop-culture inspired acronym for ‘POK erythroid initiation and progression. In prostate cancer, aberrant myeloid ontogenic factor’, is a POK protein family epigenetic events such as DNA hypo- and hypermethy- member encoded by the ZbTb7 gene and characterized lation and altered histone acetylation have been by its critical transcription function in cellular differ- observed to affect a large number of genes. However, entiation. Takahiro Maeda, Pier Paolo Pandolfi and no plenitude is without peril: although the number of colleagues show that Pokemon acts as a genuine proto- genes that undergo aberrant epigenetic inactivation oncogene when overexpressed by inhibiting the expres- associated with prostate cancer seems to be growing, sion of tumor suppressors, such as ARF. very few of these genes have yielded promising results To establish this novel finding, the authors conducted as potential tumor biomarkers for early diagnosis and a complete battery of genetic, clinical, and pathological risk assessment. -

This Thesis Has Been Submitted in Fulfilment of the Requirements for a Postgraduate Degree (E.G
This thesis has been submitted in fulfilment of the requirements for a postgraduate degree (e.g. PhD, MPhil, DClinPsychol) at the University of Edinburgh. Please note the following terms and conditions of use: • This work is protected by copyright and other intellectual property rights, which are retained by the thesis author, unless otherwise stated. • A copy can be downloaded for personal non-commercial research or study, without prior permission or charge. • This thesis cannot be reproduced or quoted extensively from without first obtaining permission in writing from the author. • The content must not be changed in any way or sold commercially in any format or medium without the formal permission of the author. • When referring to this work, full bibliographic details including the author, title, awarding institution and date of the thesis must be given. Regulation and Function of miR-199-3p in Murine and Human Cytomegalovirus Infections Nouf N. M. Laqtom s0898296 A thesis submitted in fulfilment of requirements for the degree of Doctor of Philosophy Division of Pathway Medicine, School of Biomedical Sciences The University of Edinburgh May 2013 Supervisors: Dr. Amy Buck and Dr. Bernadette Dutia i Declaration of Authorship I hereby declare that this thesis is of my own composition, and that it contains no material previously submitted for the award of any other degree. The work reported in this thesis has been executed by myself, except where due acknowledgement is made in the text. Nouf N. M. Laqtom ii Abstract Human Cytomegalovirus (HCMV), the prototypic β-herpesvirus, is the most common cause of congenital infections as well as morbidity and mortality in immunocompromised patients. -

Analysis of Gene Expression in the Nervous System Identifies Key Genes and Novel Candidates for Health and Disease
Neurogenetics (2017) 18:81–95 DOI 10.1007/s10048-017-0509-5 ORIGINAL ARTICLE Analysis of gene expression in the nervous system identifies key genes and novel candidates for health and disease Sarah M Carpanini1 & Thomas M Wishart1 & Thomas H Gillingwater2 & Jean C Manson1 & Kim M Summers1 Received: 13 July 2016 /Accepted: 20 January 2017 /Published online: 11 February 2017 # The Author(s) 2017. This article is published with open access at Springerlink.com Abstract The incidence of neurodegenerative diseases in the pathways and gene ontology term annotation. Additionally a developed world has risen over the last century, concomitant number of poorly annotated genes were implicated by this with an increase in average human lifespan. A major chal- approach in nervous system function. Exploiting gene expres- lenge is therefore to identify genes that control neuronal health sion data available in the public domain allowed us to validate and viability with a view to enhancing neuronal health during key nervous system genes and, importantly, to identify additional ageing and reducing the burden of neurodegeneration. genes with minimal functional annotation but with the same Analysis of gene expression data has recently been used to expression pattern. These genes are thus novel candidates for infer gene functions for a range of tissues from co-expression a role in neurological health and disease and could now be networks. We have now applied this approach to further investigated to confirm their function and regulation transcriptomic datasets from the mammalian nervous system during ageing and neurodegeneration. available in the public domain. We have defined the genes critical for influencing neuronal health and disease in different Keywords Mice . -

A Genomic Approach to Delineating the Occurrence of Scoliosis in Arthrogryposis Multiplex Congenita
G C A T T A C G G C A T genes Article A Genomic Approach to Delineating the Occurrence of Scoliosis in Arthrogryposis Multiplex Congenita Xenia Latypova 1, Stefan Giovanni Creadore 2, Noémi Dahan-Oliel 3,4, Anxhela Gjyshi Gustafson 2, Steven Wei-Hung Hwang 5, Tanya Bedard 6, Kamran Shazand 2, Harold J. P. van Bosse 5 , Philip F. Giampietro 7,* and Klaus Dieterich 8,* 1 Grenoble Institut Neurosciences, Université Grenoble Alpes, Inserm, U1216, CHU Grenoble Alpes, 38000 Grenoble, France; [email protected] 2 Shriners Hospitals for Children Headquarters, Tampa, FL 33607, USA; [email protected] (S.G.C.); [email protected] (A.G.G.); [email protected] (K.S.) 3 Shriners Hospitals for Children, Montreal, QC H4A 0A9, Canada; [email protected] 4 School of Physical & Occupational Therapy, Faculty of Medicine and Health Sciences, McGill University, Montreal, QC H3G 2M1, Canada 5 Shriners Hospitals for Children, Philadelphia, PA 19140, USA; [email protected] (S.W.-H.H.); [email protected] (H.J.P.v.B.) 6 Alberta Congenital Anomalies Surveillance System, Alberta Health Services, Edmonton, AB T5J 3E4, Canada; [email protected] 7 Department of Pediatrics, University of Illinois-Chicago, Chicago, IL 60607, USA 8 Institut of Advanced Biosciences, Université Grenoble Alpes, Inserm, U1209, CHU Grenoble Alpes, 38000 Grenoble, France * Correspondence: [email protected] (P.F.G.); [email protected] (K.D.) Citation: Latypova, X.; Creadore, S.G.; Dahan-Oliel, N.; Gustafson, Abstract: Arthrogryposis multiplex congenita (AMC) describes a group of conditions characterized A.G.; Wei-Hung Hwang, S.; Bedard, by the presence of non-progressive congenital contractures in multiple body areas. -

Case Report: Exome Sequencing Achieved a Definite Diagnosis in A
Jiang et al. BMC Neurology (2021) 21:96 https://doi.org/10.1186/s12883-021-02093-z CASE REPORT Open Access Case report: exome sequencing achieved a definite diagnosis in a Chinese family with muscle atrophy Hui Jiang1,2,3†, Chunmiao Guo4†, Jie Xie1,2,3†, Jingxin Pan5, Ying Huang1,2,3, Miaoxin Li1,2,3,6,7* and Yibin Guo2,3,8* Abstract Background: Due to large genetic and phenotypic heterogeneity, the conventional workup for Charcot-Marie- Tooth (CMT) diagnosis is often underpowered, leading to diagnostic delay or even lack of diagnosis. In the present study, we explored how bioinformatics analysis on whole-exome sequencing (WES) data can be used to diagnose patients with CMT disease efficiently. Case presentation: The proband is a 29-year-old female presented with a severe amyotrophy and distal skeletal deformity that plagued her family for over 20 years since she was 5-year-old. No other aberrant symptoms were detected in her speaking, hearing, vision, and intelligence. Similar symptoms manifested in her younger brother, while her parents and her older brother showed normal. To uncover the genetic causes of this disease, we performed exome sequencing for the proband and her parents. Subsequent bioinformatics analysis on the KGGSeq platform and further Sanger sequencing identified a novel homozygous GDAP1 nonsense mutation (c.218C > G, p.Ser73*) that responsible for the family. This genetic finding then led to a quick diagnosis of CMT type 4A (CMT4A), confirmed by nerve conduction velocity and electromyography examination of the patients. Conclusions: The patients with severe muscle atrophy and distal skeletal deformity were caused by a novel homozygous nonsense mutation in GDAP1 (c.218C > G, p.Ser73*), and were diagnosed as CMT4A finally. -

Global Patterns of Changes in the Gene Expression Associated with Genesis of Cancer a Dissertation Submitted in Partial Fulfillm
Global Patterns Of Changes In The Gene Expression Associated With Genesis Of Cancer A dissertation submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy at George Mason University By Ganiraju Manyam Master of Science IIIT-Hyderabad, 2004 Bachelor of Engineering Bharatiar University, 2002 Director: Dr. Ancha Baranova, Associate Professor Department of Molecular & Microbiology Fall Semester 2009 George Mason University Fairfax, VA Copyright: 2009 Ganiraju Manyam All Rights Reserved ii DEDICATION To my parents Pattabhi Ramanna and Veera Venkata Satyavathi who introduced me to the joy of learning. To friends, family and colleagues who have contributed in work, thought, and support to this project. iii ACKNOWLEDGEMENTS I would like to thank my advisor, Dr. Ancha Baranova, whose tolerance, patience, guidance and encouragement helped me throughout the study. This dissertation would not have been possible without her ever ending support. She is very sincere and generous with her knowledge, availability, compassion, wisdom and feedback. I would also like to thank Dr. Vikas Chandhoke for funding my research generously during my doctoral study at George Mason University. Special thanks go to Dr. Patrick Gillevet, Dr. Alessandro Giuliani, Dr. Maria Stepanova who devoted their time to provide me with their valuable contributions and guidance to formulate this project. Thanks to the faculty of Molecular and Micro Biology (MMB) department, Dr. Jim Willett and Dr. Monique Vanhoek in embedding valuable thoughts to this dissertation by being in my dissertation committee. I would also like to thank the present and previous doctoral program directors, Dr. Daniel Cox and Dr. Geraldine Grant, for facilitating, allowing, and encouraging me to work in this project.