Supplementary Materials Functional Characterization of Rare RAB12
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

ZMYND10 Functions in a Chaperone Relay During Axonemal Dynein Assembly
bioRxiv preprint doi: https://doi.org/10.1101/233718; this version posted December 13, 2017. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 1 ZMYND10 functions in a chaperone relay during axonemal dynein assembly. 2 3 Girish R Mali1,9 , Patricia Yeyati1, Seiya Mizuno2, Margaret A Keighren1, Petra zur Lage3, Amaya 4 Garcia-Munoz4, Atsuko Shimada5, Hiroyuki Takeda5, Frank Edlich6, Satoru Takahashi2,7, Alex von 5 Kreigsheim4,8, Andrew Jarman3 and Pleasantine Mill1,*. 6 7 1. MRC Human Genetics Unit, Institute of Genetics and Molecular Medicine, University of 8 Edinburgh, Edinburgh, UK, EH4 2XU 9 2. Laboratory Animal Resource Centre, University of Tsukuba, Tsukuba, Japan, 305-8575 10 3. Centre for Integrative Physiology, University of Edinburgh, Edinburgh, UK, EH8 9XD 11 4. Systems Biology Ireland, University College Dublin, Dublin, Ireland 12 5. Department of Biological Sciences, University of Tokyo, Tokyo, Japan, 113-0033 13 6. Institute for Biochemistry and Molecular Biology, University of Freiburg, Freiburg, Germany, 14 79104 15 7. Department of Anatomy and Embryology, Faculty of Medicine, University of Tsukuba, Tsukuba, 16 Japan, 305-8575 17 8. Edinburgh Cancer Research UK Centre, Institute of Genetics and Molecular Medicine, University 18 of Edinburgh, Edinburgh, UK, EH4 2XU 19 9. Current address: MRC Laboratory of Molecular Biology, Cambridge, UK, CB2 0QH 20 * Corresponding author: [email protected] 21 22 23 24 25 26 27 28 29 30 31 1 bioRxiv preprint doi: https://doi.org/10.1101/233718; this version posted December 13, 2017. -

Establishment of the Early Cilia Preassembly Protein Complex
Establishment of the early cilia preassembly protein PNAS PLUS complex during motile ciliogenesis Amjad Horania,1, Alessandro Ustioneb, Tao Huangc, Amy L. Firthd, Jiehong Panc, Sean P. Gunstenc, Jeffrey A. Haspelc, David W. Pistonb, and Steven L. Brodyc aDepartment of Pediatrics, Washington University School of Medicine, St. Louis, MO 63110; bDepartment of Cell Biology and Physiology, Washington University School of Medicine, St. Louis, MO 63110; cDepartment of Medicine, Washington University School of Medicine, St. Louis, MO 63110; and dDepartment of Medicine, University of Southern California, Keck School of Medicine, Los Angeles, CA 90033 Edited by Kathryn V. Anderson, Sloan Kettering Institute, New York, NY, and approved December 27, 2017 (received for review September 9, 2017) Motile cilia are characterized by dynein motor units, which preas- function of these proteins is unknown; however, missing dynein semble in the cytoplasm before trafficking into the cilia. Proteins motor complexes in the cilia of mutants and cytoplasmic locali- required for dynein preassembly were discovered by finding human zation (or absence in the cilia proteome) suggest a role in the mutations that result in absent ciliary motors, but little is known preassembly of dynein motor complexes. Studies in C. reinhardtii about their expression, function, or interactions. By monitoring show motor components in the cell body before transport to ciliogenesis in primary airway epithelial cells and MCIDAS-regulated flagella (22–25). However, the expression, interactions, and induced pluripotent stem cells, we uncovered two phases of expres- functions of preassembly proteins, as well as the steps required sion of preassembly proteins. An early phase, composed of HEATR2, for preassembly, are undefined. -

De Novo, Systemic, Deleterious Amino Acid Substitutions Are Common in Large Cytoskeleton‑Related Protein Coding Regions
BIOMEDICAL REPORTS 6: 211-216, 2017 De novo, systemic, deleterious amino acid substitutions are common in large cytoskeleton‑related protein coding regions REBECCA J. STOLL1, GRACE R. THOMPSON1, MOHAMMAD D. SAMY1 and GEORGE BLANCK1,2 1Department of Molecular Medicine, Morsani College of Medicine, University of South Florida; 2Immunology Program, H. Lee Moffitt Cancer Center and Research Institute, Tampa, FL 33612, USA Received June 13, 2016; Accepted October 31, 2016 DOI: 10.3892/br.2016.826 Abstract. Human mutagenesis is largely random, thus large Introduction coding regions, simply on the basis of probability, represent relatively large mutagenesis targets. Thus, we considered Genetic damage is largely random and therefore tends to the possibility that large cytoskeletal-protein related coding affect the larger, functional regions of the human genome regions (CPCRs), including extra-cellular matrix (ECM) more frequently than the smaller regions (1). For example, coding regions, would have systemic nucleotide variants that a systematic study has revealed that cancer fusion genes, on are not present in common SNP databases. Presumably, such average, are statistically, significantly larger than other human variants arose recently in development or in recent, preceding genes (2,3). The large introns of potential cancer fusion genes generations. Using matched breast cancer and blood-derived presumably allow for many different productive recombina- normal datasets from the cancer genome atlas, CPCR single tion opportunities, i.e., many recombinations that would allow nucleotide variants (SNVs) not present in the All SNPs(142) for exon juxtaposition and the generation of hybrid proteins. or 1000 Genomes databases were identified. Using the Protein Smaller cancer fusion genes tend to be associated with the rare Variation Effect Analyzer internet-based tool, it was discov- types of cancer, for example EWS RNA binding protein 1 in ered that apparent, systemic mutations (not shared among Ewing's sarcoma. -

Next Generation Massively Parallel Sequencing of Targeted
ARTICLE Next generation massively parallel sequencing of targeted exomes to identify genetic mutations in primary ciliary dyskinesia: Implications for application to clinical testing Jonathan S. Berg, MD, PhD1,2, James P. Evans, MD, PhD1,2, Margaret W. Leigh, MD3, Heymut Omran, MD4, Chris Bizon, PhD5, Ketan Mane, PhD5, Michael R. Knowles, MD2, Karen E. Weck, MD1,6, and Maimoona A. Zariwala, PhD6 Purpose: Advances in genetic sequencing technology have the potential rimary ciliary dyskinesia (PCD) is an autosomal recessive to enhance testing for genes associated with genetically heterogeneous Pdisorder involving abnormalities of motile cilia, resulting in clinical syndromes, such as primary ciliary dyskinesia. The objective of a range of manifestations including situs inversus, neonatal this study was to investigate the performance characteristics of exon- respiratory distress at full-term birth, recurrent otitis media, capture technology coupled with massively parallel sequencing for chronic sinusitis, chronic bronchitis that may result in bronchi- 1–3 clinical diagnostic evaluation. Methods: We performed a pilot study of ectasis, and male infertility. The disorder is genetically het- four individuals with a variety of previously identified primary ciliary erogeneous, rendering molecular diagnosis challenging given dyskinesia mutations. We designed a custom array (NimbleGen) to that mutations in nine different genes (DNAH5, DNAH11, capture 2089 exons from 79 genes associated with primary ciliary DNAI1, DNAI2, KTU, LRRC50, RSPH9, RSPH4A, and TX- dyskinesia or ciliary function and sequenced the enriched material using NDC3) account for only approximately 1/3 of PCD cases.4 the GS FLX Titanium (Roche 454) platform. Bioinformatics analysis DNAH5 and DNAI1 account for the majority of known muta- was performed in a blinded fashion in an attempt to detect the previ- tions, and the other genes each account for a small number of ously identified mutations and validate the process. -

Antibody List 2014
Antibody List 2014 Office of Technology Transfer, DKFZ Terms and Conditions Terms and conditions for licensing DKFZ’s antibodies are available upon request. Disclaimer This antibody list is neither exhaustive nor qualifies as a commercial catalogue. DKFZ has prepared the contents of this list to the best of its knowledge and confirms to the best of its knowledge that the listed antibodies are created at or in cooperation with DKFZ. DKFZ does not warrant that any listed antibody is available. Moreover DKFZ does not warrant that any listed antibody and / or the use thereof is not protected by any third party rights. Contact: Office of Technology Transfer (T010) Deutsches Krebsforschungszentrum (DKFZ) German Cancer Research Center Im Neuenheimer Feld 280 69120 Heidelberg Germany E-mail: [email protected] Index MMP9 AATF Hexa-His MPO ABCB5 HIP128 Myo II AID HIP150 NET1 AGO1 HIPK-2 Nephrin AuroraC HIPPI NF1 APC Histone H1.0 NM-1 (NMy1) BPAG Histone H1.2 NMUR-2 BPV1 L1 Histone H1.3 NO-52 BPV1 E7 Histone H1.4 NO-66 BTG2 HLA-DM NO-145 CCDC103 HLA-DO NPHP1 CCR4 HLA-DP NPHP3 CCR8 HLA-DQ NPHP4 CCR9 HLA-DR NPM CCR10 HPV11 L1 Nup88 CD3 HPV11 E2 Nus/S-tag CD19 HPV16 L1 OCAB CD22 HPV16 L2 ODA7 CD23 HPV16 E5 PACS-1 CD37 HPV16 E6 PACT CD53 HPV16 E7 p16 CD56 HPV18 E6 p27KIP CD71 HPV18 E7 p53 CD171 (L1CAM) HPV20 E6 Pick CEACAM1 HPV20 E7 PKD1 Cep170 HPV27 E6 PIF Densin HPV27 E7 Plasminogen acitvator (PA) DLL4 HSC70 Podocin DNAH3 HSP70 PSA DNAH5 Jade1 PSMA DNAH8 JAM4 PTEN DNAH11 Ki67 RFP2 DNAse1 KTU RGS7 DSC2 LaminA RIP140 EGFR LaminB1 -

Supplementary Table S4. FGA Co-Expressed Gene List in LUAD
Supplementary Table S4. FGA co-expressed gene list in LUAD tumors Symbol R Locus Description FGG 0.919 4q28 fibrinogen gamma chain FGL1 0.635 8p22 fibrinogen-like 1 SLC7A2 0.536 8p22 solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 DUSP4 0.521 8p12-p11 dual specificity phosphatase 4 HAL 0.51 12q22-q24.1histidine ammonia-lyase PDE4D 0.499 5q12 phosphodiesterase 4D, cAMP-specific FURIN 0.497 15q26.1 furin (paired basic amino acid cleaving enzyme) CPS1 0.49 2q35 carbamoyl-phosphate synthase 1, mitochondrial TESC 0.478 12q24.22 tescalcin INHA 0.465 2q35 inhibin, alpha S100P 0.461 4p16 S100 calcium binding protein P VPS37A 0.447 8p22 vacuolar protein sorting 37 homolog A (S. cerevisiae) SLC16A14 0.447 2q36.3 solute carrier family 16, member 14 PPARGC1A 0.443 4p15.1 peroxisome proliferator-activated receptor gamma, coactivator 1 alpha SIK1 0.435 21q22.3 salt-inducible kinase 1 IRS2 0.434 13q34 insulin receptor substrate 2 RND1 0.433 12q12 Rho family GTPase 1 HGD 0.433 3q13.33 homogentisate 1,2-dioxygenase PTP4A1 0.432 6q12 protein tyrosine phosphatase type IVA, member 1 C8orf4 0.428 8p11.2 chromosome 8 open reading frame 4 DDC 0.427 7p12.2 dopa decarboxylase (aromatic L-amino acid decarboxylase) TACC2 0.427 10q26 transforming, acidic coiled-coil containing protein 2 MUC13 0.422 3q21.2 mucin 13, cell surface associated C5 0.412 9q33-q34 complement component 5 NR4A2 0.412 2q22-q23 nuclear receptor subfamily 4, group A, member 2 EYS 0.411 6q12 eyes shut homolog (Drosophila) GPX2 0.406 14q24.1 glutathione peroxidase -
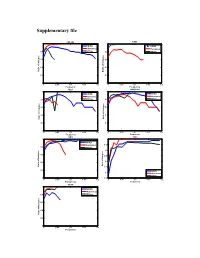
Supplementary File
Supplementary file COL1A2 FZD3 1 1 HPRD HPRD Humannet Humannet 0.8 PPIwu 0.8 PPIwu 0.6 0.6 0.4 0.4 Ratio of of indegree Ratio Ratio of Indegree 0.2 0.2 0 0 0 0.05 0.1 0.15 0.2 0 0.05 0.1 0.15 0.2 Frequency Frequency RBL1 SMARCA4 1 1 HPRD HPRD Humannet Humannet 0.8 PPIwu 0.8 PPIwu 0.6 0.6 0.4 0.4 Ratio of Indegree Ratio of Indegree Ratio 0.2 0.2 0 0 0 0.05 0.1 0.15 0.2 0 0.05 0.1 0.15 0.2 Frequency Frequency SRC TP53 1 1 HPRD Humannet 0.95 0.8 PPIwu 0.9 e 0.6 0.85 0.8 0.4 Ratio of Indegree of Ratio Ratio of Indegre 0.75 0.2 HPRD 0.7 Humannet PPIwu 0 0.65 0 0.05 0.1 0.15 0.2 0 0.05 0.1 0.15 0.2 Frequency Frequency VCAN 1 HPRD Humannet 0.8 PPIwu 0.6 0.4 Ratio of Indegree 0.2 0 0 0.05 0.1 0.15 0.2 Frequency Figure S1. Variation of indegree ratio with frequency cutoffs from 0 to 0.2 with a step 0.002 for driver genes common by three fitness cores in COAD. The indegree ratio is set to NULL if total degree of this genes is less than 5. BRAF CASR 1 1 0.8 0.8 0.6 0.6 0.4 0.4 Ratio of Indegree Ratio of Indegree of Ratio HPRD HPRD 0.2 0.2 Humannet Humannet PPIwu PPIwu 0 0 0 0.05 0.1 0.15 0.2 0 0.05 0.1 0.15 0.2 Frequency Frequency HDAC9 NRAS 1 1 0.8 0.8 0.6 0.6 0.4 0.4 HPRD HPRD Ratio of Indegree Ratio of Indegree 0.2 0.2 Humannet Humannet PPIwu PPIwu 0 0 0 0.05 0.1 0.15 0.2 0 0.05 0.1 0.15 0.2 Frequency Frequency NF1 CNTNAP2 1 1 0.9 0.8 0.8 0.6 0.7 0.4 HPRD HPRD Ratio of Indegree Ratio of Indegree 0.6 Humannet 0.2 Humannet PPIwu PPIwu 0.5 0 0 0.05 0.1 0.15 0.2 0 0.05 0.1 0.15 0.2 Frequency Frequency Figure S2. -

Identification of Tumor Mutation Burden-Related Hub Genes and the Underlying Mechanism in Melanoma
Journal of Cancer 2021, Vol. 12 2440 Ivyspring International Publisher Journal of Cancer 2021; 12(8): 2440-2449. doi: 10.7150/jca.53697 Research Paper Identification of tumor mutation burden-related hub genes and the underlying mechanism in melanoma Chuan Zhang1,2, Dan Dang3, Chenlu Liu4, Yuqian Wang5, Xianling Cong1 1. Department of Dermatology, China-Japan Union Hospital of Jilin University, Changchun 130033, People’s Republic of China. 2. Department of Pediatric Surgery, the First Hospital of Jilin University, Changchun 130021, People’s Republic of China. 3. Department of Neonatology, the First Hospital of Jilin University, Changchun 130021, People’s Republic of China. 4. Department of Tissue Bank, China-Japan Union Hospital of Jilin University, Changchun, 130033, People’s Republic of China. 5. Scientific Research Center, China-Japan Union Hospital of Jilin University, Changchun 130033, People’s Republic of China. Corresponding author: Xianling Cong M.D. (E-mail: [email protected]), Department of Dermatology, China-Japan Union Hospital of Jilin University, Changchun 130033, People’s Republic of China, 130033. © The author(s). This is an open access article distributed under the terms of the Creative Commons Attribution License (https://creativecommons.org/licenses/by/4.0/). See http://ivyspring.com/terms for full terms and conditions. Received: 2020.09.24; Accepted: 2021.01.25; Published: 2021.03.01 Abstract Background: Tumor mutation burden (TMB) has emerged as an important predictive factor for drug resistance in cancers; however, the specific mechanism underlying TMB function in melanoma remains elusive. Methods: Data on somatic mutations, RNA sequencing (RNA-seq), miRNA sequencing (miRNA-seq), and clinical characteristics for 472 melanoma patients were extracted from the TCGA cohort. -

Integrated Next Generation Sequencing and Avatar Mouse Models for Personalized Cancer Treatment Elena Garralda1, Keren Paz2
Author Manuscript Published OnlineFirst on March 14, 2014; DOI: 10.1158/1078-0432.CCR-13-3047 Author manuscripts have been peer reviewed and accepted for publication but have not yet been edited. 1 Integrated Next Generation Sequencing and Avatar Mouse Models for Personalized Cancer Treatment Elena Garralda1, Keren Paz2, Pedro P. López-Casas1, Siân Jones3, Amanda Katz2, Lisa M. Kann3, Fernando López-Rios4, Francesca Sarno5, Fátima Al- Shahrour1, David Vasquez2, Elizabeth Bruckheimer2, Samuel V. Angiuoli3, Antonio Calles1, Luis A. Diaz6, Victor E. Velculescu6, Alfonso Valencia1, David Sidransky2, Manuel Hidalgo1. 1Spanish National Cancer Research Centre (CNIO), Madrid, Spain; 2Champions Oncology, Baltimore, MD; 3Personal Genome Diagnostics, Inc., Baltimore, MD; 4Laboratorio Dianas Terapéuticas, Hospital Universitario Madrid-Sanchinarro, Madrid, Spain; 5Centro Integral Oncológico Clara Campal, Hospital Universitario Madrid-Sanchinarro, Madrid, Spain; 6Ludwig Center for Cancer Genetics and Therapeutics, Johns Hopkins Kimmel Cancer Center, Baltimore, MD Running Title: Personalized Cancer Treatment using Genomics and Avatar Models Key Words: Personalized Medicine, Next-Generation Sequencing, Avatar Mouse Models Word count: 3419 Tables: 3 Figures: 2 Correspondence: • Manuel Hidalgo, [email protected] Downloaded from clincancerres.aacrjournals.org on September 27, 2021. © 2014 American Association for Cancer Research. Author Manuscript Published OnlineFirst on March 14, 2014; DOI: 10.1158/1078-0432.CCR-13-3047 Author manuscripts have been peer reviewed and accepted for publication but have not yet been edited. 2 Centro Nacional de Investigaciones Oncológicas (CNIO) (Spanish National Cancer Research Centre) Melchor Fernández Almagro nº 3, E-28029 Madrid, Spain Tel.: Tel.: +34 91 732 80 00 Ext. 2920 Fax +34 915360432 Conflict of interest: L.A.D., and V.E.V. -

Supplementary Material Contents
Supplementary Material Contents Immune modulating proteins identified from exosomal samples.....................................................................2 Figure S1: Overlap between exosomal and soluble proteomes.................................................................................... 4 Bacterial strains:..............................................................................................................................................4 Figure S2: Variability between subjects of effects of exosomes on BL21-lux growth.................................................... 5 Figure S3: Early effects of exosomes on growth of BL21 E. coli .................................................................................... 5 Figure S4: Exosomal Lysis............................................................................................................................................ 6 Figure S5: Effect of pH on exosomal action.................................................................................................................. 7 Figure S6: Effect of exosomes on growth of UPEC (pH = 6.5) suspended in exosome-depleted urine supernatant ....... 8 Effective exosomal concentration....................................................................................................................8 Figure S7: Sample constitution for luminometry experiments..................................................................................... 8 Figure S8: Determining effective concentration ......................................................................................................... -

Supplementary Table 1. Mutated Genes That Contain Protein Domains Identified Through Mutation Enrichment Analysis
Supplementary Table 1. Mutated genes that contain protein domains identified through mutation enrichment analysis A. Breast cancers InterPro ID Mutated genes (number of mutations) IPR000219 ARHGEF4(2), ECT2(1), FARP1(1), FLJ20184(1), MCF2L2(1), NET1(1), OBSCN(5), RASGRF2(2), TRAD(1), VAV3(1) IPR000225 APC2(2), JUP(1), KPNA5(2), SPAG6(1) IPR000357 ARFGEF2(2), CMYA4(1), DRIM(2), JUP(1), KPNA5(2), PIK3R4(1), SPAG6(1) IPR000533 AKAP9(2), C10orf39(1), C20orf23(1), CUTL1(1), HOOK1(1), HOOK3(1), KTN1(2), LRRFIP1(3), MYH1(3), MYH9(2), NEF3(1), NF2(1), RSN(1), TAX1BP1(1), TPM4(1) IPR000694 ADAM12(3), ADAMTS19(1), APC2(2), APXL(1), ARID1B(1), BAT2(2), BAT3(1), BCAR1(1), BCL11A(2), BCORL1(1), C14orf155(3), C1orf2(1), C1QB(1), C6orf31(1), C7orf11(1), CD2(1), CENTD3(3), CHD5(3), CIC(3), CMYA1(2), COL11A1(3), COL19A1(2), COL7A1(3), DAZAP1(1), DBN1(3), DVL3(1), EIF5(1), FAM44A(1), FAM47B(1), FHOD1(1), FLJ20584(1), G3BP2(2), GAB1(2), GGA3(1), GLI1(3), GPNMB(2), GRIN2D(3), HCN3(1), HOXA3(2), HOXA4(1), IRS4(1), KCNA5(1), KCNC2(1), LIP8(1), LOC374955(1), MAGEE1(2), MICAL1(2), MICAL‐L1(1), MLLT2(1), MMP15(1), N4BP2(1), NCOA6(2), NHS(1), NUP214(3), ODZ1(3), PER1(2), PER2(1), PHC1(1), PLXNB1(1), PPM1E(2), RAI17(2), RAPH1(2), RBAF600(2), SCARF2(1), SEMA4G(1), SLC16A2(1), SORBS1(1), SPEN(2), SPG4(1), TBX1(1), TCF1(2), TCF7L1(1), TESK1(1), THG‐1(1), TP53(18), TRIF(1), ZBTB3(2), ZNF318(2) IPR000909 CENTB1(2), PLCB1(1), PLCG1(1) IPR000998 AEGP(3), EGFL6(2), PRSS7(1) IPR001140 ABCB10(2), ABCB6(1), ABCB8(2) IPR001164 ARFGAP3(1), CENTB1(2), CENTD3(3), CENTG1(2) IPR001589 -

Novel Targets of Apparently Idiopathic Male Infertility
International Journal of Molecular Sciences Review Molecular Biology of Spermatogenesis: Novel Targets of Apparently Idiopathic Male Infertility Rossella Cannarella * , Rosita A. Condorelli , Laura M. Mongioì, Sandro La Vignera * and Aldo E. Calogero Department of Clinical and Experimental Medicine, University of Catania, 95123 Catania, Italy; [email protected] (R.A.C.); [email protected] (L.M.M.); [email protected] (A.E.C.) * Correspondence: [email protected] (R.C.); [email protected] (S.L.V.) Received: 8 February 2020; Accepted: 2 March 2020; Published: 3 March 2020 Abstract: Male infertility affects half of infertile couples and, currently, a relevant percentage of cases of male infertility is considered as idiopathic. Although the male contribution to human fertilization has traditionally been restricted to sperm DNA, current evidence suggest that a relevant number of sperm transcripts and proteins are involved in acrosome reactions, sperm-oocyte fusion and, once released into the oocyte, embryo growth and development. The aim of this review is to provide updated and comprehensive insight into the molecular biology of spermatogenesis, including evidence on spermatogenetic failure and underlining the role of the sperm-carried molecular factors involved in oocyte fertilization and embryo growth. This represents the first step in the identification of new possible diagnostic and, possibly, therapeutic markers in the field of apparently idiopathic male infertility. Keywords: spermatogenetic failure; embryo growth; male infertility; spermatogenesis; recurrent pregnancy loss; sperm proteome; DNA fragmentation; sperm transcriptome 1. Introduction Infertility is a widespread condition in industrialized countries, affecting up to 15% of couples of childbearing age [1]. It is defined as the inability to achieve conception after 1–2 years of unprotected sexual intercourse [2].