From Syndromes to Normal Variation: a Candidate Gene Study of Interorbital Distances
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Tall Stature: a Difficult Diagnosis? Cristina Meazza1* , Chiara Gertosio2, Roberta Giacchero3, Sara Pagani1 and Mauro Bozzola1
Meazza et al. Italian Journal of Pediatrics (2017) 43:66 DOI 10.1186/s13052-017-0385-5 REVIEW Open Access Tall stature: a difficult diagnosis? Cristina Meazza1* , Chiara Gertosio2, Roberta Giacchero3, Sara Pagani1 and Mauro Bozzola1 Abstract Referral for an assessment of tall stature is less common than for short stature. Tall stature is defined as a height more than two standard deviations above the mean for age. The majority of subjects with tall stature show a familial tall stature or a constitutional advance of growth (CAG), which is a diagnosis of exclusion. After a careful physical evaluation, tall subjects may be divided into two groups: tall subjects with normal appearance and tall subjects with abnormal appearance. In the case of normal appearance, the paediatric endocrinologist will have to evaluate the growth rate. If it is normal for age and sex, the subject may be classified as having familial tall stature, CAG or obese subject, while if the growth rate is increased it is essential to evaluate pubertal status and thyroid status. Tall subjects with abnormal appearance and dysmorphisms can be classified into those with proportionate and disproportionate syndromes. A careful physical examination and an evaluation of growth pattern are required before starting further investigations. Physicians should always search for a pathological cause of tall stature, although the majority of children are healthy and they generally do not need treatment to cease growth progression. The most accepted and effective treatment for an excessive height prediction is inducing puberty early and leading to a complete fusion of the epiphyses and achievement of final height, using testosterone in males and oestrogens in females. -

Identification of the Binding Partners for Hspb2 and Cryab Reveals
Brigham Young University BYU ScholarsArchive Theses and Dissertations 2013-12-12 Identification of the Binding arP tners for HspB2 and CryAB Reveals Myofibril and Mitochondrial Protein Interactions and Non- Redundant Roles for Small Heat Shock Proteins Kelsey Murphey Langston Brigham Young University - Provo Follow this and additional works at: https://scholarsarchive.byu.edu/etd Part of the Microbiology Commons BYU ScholarsArchive Citation Langston, Kelsey Murphey, "Identification of the Binding Partners for HspB2 and CryAB Reveals Myofibril and Mitochondrial Protein Interactions and Non-Redundant Roles for Small Heat Shock Proteins" (2013). Theses and Dissertations. 3822. https://scholarsarchive.byu.edu/etd/3822 This Thesis is brought to you for free and open access by BYU ScholarsArchive. It has been accepted for inclusion in Theses and Dissertations by an authorized administrator of BYU ScholarsArchive. For more information, please contact [email protected], [email protected]. Identification of the Binding Partners for HspB2 and CryAB Reveals Myofibril and Mitochondrial Protein Interactions and Non-Redundant Roles for Small Heat Shock Proteins Kelsey Langston A thesis submitted to the faculty of Brigham Young University in partial fulfillment of the requirements for the degree of Master of Science Julianne H. Grose, Chair William R. McCleary Brian Poole Department of Microbiology and Molecular Biology Brigham Young University December 2013 Copyright © 2013 Kelsey Langston All Rights Reserved ABSTRACT Identification of the Binding Partners for HspB2 and CryAB Reveals Myofibril and Mitochondrial Protein Interactors and Non-Redundant Roles for Small Heat Shock Proteins Kelsey Langston Department of Microbiology and Molecular Biology, BYU Master of Science Small Heat Shock Proteins (sHSP) are molecular chaperones that play protective roles in cell survival and have been shown to possess chaperone activity. -

Genetic Investigation of Patients with Tall Stature
2 182 E Vasco de Albuquerque Genetic investigation of tall 182:2 139–147 Clinical Study Albuquerque and others stature patients Genetic investigation of patients with tall stature Edoarda Vasco de Albuquerque Albuquerque1, Mariana Ferreira de Assis Funari2, Elisângela Pereira de Souza Quedas1, Rachel Sayuri Honjo Kawahira3, Raquel Soares Jallad4, Thaís Kataoka Homma1, Regina Matsunaga Martin2,5, Vinicius Nahime Brito2, Alexsandra Christianne Malaquias1,6, Antonio Marcondes Lerario1,7, Carla Rosenberg8, Ana Cristina Victorino Krepischi8, Chong Ae Kim3, Ivo Jorge Prado Arnhold2 and Alexander Augusto de Lima Jorge1 1Unidade de Endocrinologia Genética (LIM25), 2Unidade de Endocrinologia do Desenvolvimento, Laboratório de Hormônios e Genética Molecular (LIM42), Hospital das Clínicas da Faculdade de Medicina, Universidade de São Paulo (USP), São Paulo, Brasil, 3Unidade de Genética do Instituto da Criança, Hospital das Clínicas da Faculdade de Medicina da Universidade de São Paulo, São Paulo, Brasil, 4Unidade de Neuroendocrinologia, 5Unidade de Doenças Osteometabólicas, Hospital das Clínicas da Faculdade de Medicina, Universidade de São Paulo (USP), São Paulo, Brasil, 6Unidade de Endocrinologia Pediátrica, Correspondence Departamento de Pediatria, Irmandade da Santa Casa de Misericórdia de São Paulo, São Paulo, Brasil, 7Division of should be addressed Metabolism, Endocrinology and Diabetes, Department of Internal Medicine, University of Michigan, Ann Arbor, to A A L Jorge Michigan, USA, and 8Instituto de Biociências (IB), Universidade de São Paulo (USP), São Paulo, Brasil Email [email protected] Abstract Context: Patients with tall stature often remain undiagnosed after clinical investigation and few studies have genetically assessed this group, most of them without a systematic approach. Objective: To assess prospectively a group of individuals with tall stature, with and without syndromic features, and to establish a molecular diagnosis for their growth disorder. -

Megalencephaly and Macrocephaly
277 Megalencephaly and Macrocephaly KellenD.Winden,MD,PhD1 Christopher J. Yuskaitis, MD, PhD1 Annapurna Poduri, MD, MPH2 1 Department of Neurology, Boston Children’s Hospital, Boston, Address for correspondence Annapurna Poduri, Epilepsy Genetics Massachusetts Program, Division of Epilepsy and Clinical Electrophysiology, 2 Epilepsy Genetics Program, Division of Epilepsy and Clinical Department of Neurology, Fegan 9, Boston Children’s Hospital, 300 Electrophysiology, Department of Neurology, Boston Children’s Longwood Avenue, Boston, MA 02115 Hospital, Boston, Massachusetts (e-mail: [email protected]). Semin Neurol 2015;35:277–287. Abstract Megalencephaly is a developmental disorder characterized by brain overgrowth secondary to increased size and/or numbers of neurons and glia. These disorders can be divided into metabolic and developmental categories based on their molecular etiologies. Metabolic megalencephalies are mostly caused by genetic defects in cellular metabolism, whereas developmental megalencephalies have recently been shown to be caused by alterations in signaling pathways that regulate neuronal replication, growth, and migration. These disorders often lead to epilepsy, developmental disabilities, and Keywords behavioral problems; specific disorders have associations with overgrowth or abnor- ► megalencephaly malities in other tissues. The molecular underpinnings of many of these disorders are ► hemimegalencephaly now understood, providing insight into how dysregulation of critical pathways leads to ► -

Supplementary Table 1: Adhesion Genes Data Set
Supplementary Table 1: Adhesion genes data set PROBE Entrez Gene ID Celera Gene ID Gene_Symbol Gene_Name 160832 1 hCG201364.3 A1BG alpha-1-B glycoprotein 223658 1 hCG201364.3 A1BG alpha-1-B glycoprotein 212988 102 hCG40040.3 ADAM10 ADAM metallopeptidase domain 10 133411 4185 hCG28232.2 ADAM11 ADAM metallopeptidase domain 11 110695 8038 hCG40937.4 ADAM12 ADAM metallopeptidase domain 12 (meltrin alpha) 195222 8038 hCG40937.4 ADAM12 ADAM metallopeptidase domain 12 (meltrin alpha) 165344 8751 hCG20021.3 ADAM15 ADAM metallopeptidase domain 15 (metargidin) 189065 6868 null ADAM17 ADAM metallopeptidase domain 17 (tumor necrosis factor, alpha, converting enzyme) 108119 8728 hCG15398.4 ADAM19 ADAM metallopeptidase domain 19 (meltrin beta) 117763 8748 hCG20675.3 ADAM20 ADAM metallopeptidase domain 20 126448 8747 hCG1785634.2 ADAM21 ADAM metallopeptidase domain 21 208981 8747 hCG1785634.2|hCG2042897 ADAM21 ADAM metallopeptidase domain 21 180903 53616 hCG17212.4 ADAM22 ADAM metallopeptidase domain 22 177272 8745 hCG1811623.1 ADAM23 ADAM metallopeptidase domain 23 102384 10863 hCG1818505.1 ADAM28 ADAM metallopeptidase domain 28 119968 11086 hCG1786734.2 ADAM29 ADAM metallopeptidase domain 29 205542 11085 hCG1997196.1 ADAM30 ADAM metallopeptidase domain 30 148417 80332 hCG39255.4 ADAM33 ADAM metallopeptidase domain 33 140492 8756 hCG1789002.2 ADAM7 ADAM metallopeptidase domain 7 122603 101 hCG1816947.1 ADAM8 ADAM metallopeptidase domain 8 183965 8754 hCG1996391 ADAM9 ADAM metallopeptidase domain 9 (meltrin gamma) 129974 27299 hCG15447.3 ADAMDEC1 ADAM-like, -

Level Estimates of Maternal Smoking and Nicotine Replacement Therapy During Pregnancy
Using primary care data to assess population- level estimates of maternal smoking and nicotine replacement therapy during pregnancy Nafeesa Nooruddin Dhalwani BSc MSc Thesis submitted to the University of Nottingham for the degree of Doctor of Philosophy November 2014 ABSTRACT Background: Smoking in pregnancy is the most significant preventable cause of poor health outcomes for women and their babies and, therefore, is a major public health concern. In the UK there is a wide range of interventions and support for pregnant women who want to quit. One of these is nicotine replacement therapy (NRT) which has been widely available for retail purchase and prescribing to pregnant women since 2005. However, measures of NRT prescribing in pregnant women are scarce. These measures are vital to assess its usefulness in smoking cessation during pregnancy at a population level. Furthermore, evidence of NRT safety in pregnancy for the mother and child’s health so far is nebulous, with existing studies being small or using retrospectively reported exposures. Aims and Objectives: The main aim of this work was to assess population- level estimates of maternal smoking and NRT prescribing in pregnancy and the safety of NRT for both the mother and the child in the UK. Currently, the only population-level data on UK maternal smoking are from repeated cross-sectional surveys or routinely collected maternity data during pregnancy or at delivery. These obtain information at one point in time, and there are no population-level data on NRT use available. As a novel approach, therefore, this thesis used the routinely collected primary care data that are currently available for approximately 6% of the UK population and provide longitudinal/prospectively recorded information throughout pregnancy. -
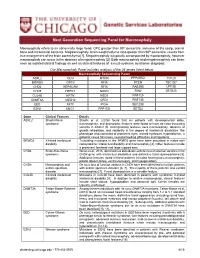
Macrocephaly Information Sheet 6-13-19
Next Generation Sequencing Panel for Macrocephaly Clinical Features: Macrocephaly refers to an abnormally large head, OFC greater than 98th percentile, inclusive of the scalp, cranial bone and intracranial contents. Megalencephaly, brain weight/volume ratio greater than 98th percentile, results from true enlargement of the brain parenchyma [1]. Megalencephaly is typically accompanied by macrocephaly, however macrocephaly can occur in the absence of megalencephaly [2]. Both macrocephaly and megalencephaly can been seen as isolated clinical findings as well as clinical features of a mutli-systemic syndromic diagnosis. Our Macrocephaly Panel includes analysis of the 36 genes listed below. Macrocephaly Sequencing Panel ASXL2 GLI3 MTOR PPP2R5D TCF20 BRWD3 GPC3 NFIA PTEN TBC1D7 CHD4 HEPACAM NFIX RAB39B UPF3B CHD8 HERC1 NONO RIN2 ZBTB20 CUL4B KPTN NSD1 RNF125 DNMT3A MED12 OFD1 RNF135 EED MITF PIGA SEC23B EZH2 MLC1 PPP1CB SETD2 Gene Clinical Features Details ASXL2 Shashi-Pena Shashi et al. (2016) found that six patients with developmental delay, syndrome macrocephaly, and dysmorphic features were found to have de novo truncating variants in ASXL2 [3]. Distinguishing features were macrocephaly, absence of growth retardation, and variability in the degree of intellectual disabilities The phenotype also consisted of prominent eyes, arched eyebrows, hypertelorism, a glabellar nevus flammeus, neonatal feeding difficulties and hypotonia. BRWD3 X-linked intellectual Truncating mutations in the BRWD3 gene have been described in males with disability nonsyndromic intellectual disability and macrocephaly [4]. Other features include a prominent forehead and large cupped ears. CHD4 Sifrim-Hitz-Weiss Weiss et al., 2016, identified five individuals with de novo missense variants in the syndrome CHD4 gene with intellectual disabilities and distinctive facial dysmorphisms [5]. -

Genome Sequences of Tropheus Moorii and Petrochromis Trewavasae, Two Eco‑Morphologically Divergent Cichlid Fshes Endemic to Lake Tanganyika C
www.nature.com/scientificreports OPEN Genome sequences of Tropheus moorii and Petrochromis trewavasae, two eco‑morphologically divergent cichlid fshes endemic to Lake Tanganyika C. Fischer1,2, S. Koblmüller1, C. Börger1, G. Michelitsch3, S. Trajanoski3, C. Schlötterer4, C. Guelly3, G. G. Thallinger2,5* & C. Sturmbauer1,5* With more than 1000 species, East African cichlid fshes represent the fastest and most species‑rich vertebrate radiation known, providing an ideal model to tackle molecular mechanisms underlying recurrent adaptive diversifcation. We add high‑quality genome reconstructions for two phylogenetic key species of a lineage that diverged about ~ 3–9 million years ago (mya), representing the earliest split of the so‑called modern haplochromines that seeded additional radiations such as those in Lake Malawi and Victoria. Along with the annotated genomes we analysed discriminating genomic features of the study species, each representing an extreme trophic morphology, one being an algae browser and the other an algae grazer. The genomes of Tropheus moorii (TM) and Petrochromis trewavasae (PT) comprise 911 and 918 Mbp with 40,300 and 39,600 predicted genes, respectively. Our DNA sequence data are based on 5 and 6 individuals of TM and PT, and the transcriptomic sequences of one individual per species and sex, respectively. Concerning variation, on average we observed 1 variant per 220 bp (interspecifc), and 1 variant per 2540 bp (PT vs PT)/1561 bp (TM vs TM) (intraspecifc). GO enrichment analysis of gene regions afected by variants revealed several candidates which may infuence phenotype modifcations related to facial and jaw morphology, such as genes belonging to the Hedgehog pathway (SHH, SMO, WNT9A) and the BMP and GLI families. -

Biallelic Alteration and Dysregulation of the Hippo Pathway in Mucinous Tubular and Spindle Cell Carcinoma of the Kidney
Published OnlineFirst September 7, 2016; DOI: 10.1158/2159-8290.CD-16-0267 RESEARCH BRIEF Biallelic Alteration and Dysregulation of the Hippo Pathway in Mucinous Tubular and Spindle Cell Carcinoma of the Kidney Rohit Mehra 1 , 2 , 3 , Pankaj Vats 1 , 3 , 4 , Marcin Cieslik 3 , Xuhong Cao 3 , 5 , Fengyun Su 3 , Sudhanshu Shukla 3 , Aaron M. Udager 1 , Rui Wang 3 , Jincheng Pan 6 , Katayoon Kasaian 3 , Robert Lonigro 3 , Javed Siddiqui 3 , Kumpati Premkumar 4 , Ganesh Palapattu 7 , Alon Weizer 2 , 7 , Khaled S. Hafez 7 , J. Stuart Wolf Jr 7 , Ankur R. Sangoi 8 , Kiril Trpkov 9 , Adeboye O. Osunkoya 10 , Ming Zhou 11 , Giovanna A. Giannico 12 , Jesse K. McKenney 13 , Saravana M. Dhanasekaran 1 , 3 , and Arul M. Chinnaiyan 1 , 2 , 3 , 5 , 7 ABSTRACT Mucinous tubular and spindle cell carcinoma (MTSCC) is a relatively rare subtype of renal cell carcinoma (RCC) with distinctive morphologic and cytogenetic fea- tures. Here, we carry out whole-exome and transcriptome sequencing of a multi-institutional cohort of MTSCC (n = 22). We demonstrate the presence of either biallelic loss of Hippo pathway tumor sup- pressor genes (TSG) and/or evidence of alteration of Hippo pathway genes in 85% of samples. PTPN14 (31%) and NF2 (22%) were the most commonly implicated Hippo pathway genes, whereas other genes such as SAV1 and HIPK2 were also involved in a mutually exclusive fashion. Mutations in the context of recurrent chromosomal losses amounted to biallelic alterations in these TSGs. As a readout of Hippo pathway inactivation, a majority of cases (90%) exhibited increased nuclear YAP1 protein expression. -

Orphanet Report Series Rare Diseases Collection
Marche des Maladies Rares – Alliance Maladies Rares Orphanet Report Series Rare Diseases collection DecemberOctober 2013 2009 List of rare diseases and synonyms Listed in alphabetical order www.orpha.net 20102206 Rare diseases listed in alphabetical order ORPHA ORPHA ORPHA Disease name Disease name Disease name Number Number Number 289157 1-alpha-hydroxylase deficiency 309127 3-hydroxyacyl-CoA dehydrogenase 228384 5q14.3 microdeletion syndrome deficiency 293948 1p21.3 microdeletion syndrome 314655 5q31.3 microdeletion syndrome 939 3-hydroxyisobutyric aciduria 1606 1p36 deletion syndrome 228415 5q35 microduplication syndrome 2616 3M syndrome 250989 1q21.1 microdeletion syndrome 96125 6p subtelomeric deletion syndrome 2616 3-M syndrome 250994 1q21.1 microduplication syndrome 251046 6p22 microdeletion syndrome 293843 3MC syndrome 250999 1q41q42 microdeletion syndrome 96125 6p25 microdeletion syndrome 6 3-methylcrotonylglycinuria 250999 1q41-q42 microdeletion syndrome 99135 6-phosphogluconate dehydrogenase 67046 3-methylglutaconic aciduria type 1 deficiency 238769 1q44 microdeletion syndrome 111 3-methylglutaconic aciduria type 2 13 6-pyruvoyl-tetrahydropterin synthase 976 2,8 dihydroxyadenine urolithiasis deficiency 67047 3-methylglutaconic aciduria type 3 869 2A syndrome 75857 6q terminal deletion 67048 3-methylglutaconic aciduria type 4 79154 2-aminoadipic 2-oxoadipic aciduria 171829 6q16 deletion syndrome 66634 3-methylglutaconic aciduria type 5 19 2-hydroxyglutaric acidemia 251056 6q25 microdeletion syndrome 352328 3-methylglutaconic -

Sotos Syndrome
European Journal of Human Genetics (2007) 15, 264–271 & 2007 Nature Publishing Group All rights reserved 1018-4813/07 $30.00 www.nature.com/ejhg PRACTICAL GENETICS In association with Sotos syndrome Sotos syndrome is an autosomal dominant condition characterised by a distinctive facial appearance, learning disability and overgrowth resulting in tall stature and macrocephaly. In 2002, Sotos syndrome was shown to be caused by mutations and deletions of NSD1, which encodes a histone methyltransferase implicated in chromatin regulation. More recently, the NSD1 mutational spectrum has been defined, the phenotype of Sotos syndrome clarified and diagnostic and management guidelines developed. Introduction In brief Sotos syndrome was first described in 1964 by Juan Sotos Sotos syndrome is characterised by a distinctive facial and the major diagnostic criteria of a distinctive facial appearance, learning disability and childhood over- appearance, childhood overgrowth and learning disability growth. were established in 1994 by Cole and Hughes.1,2 In 2002, Sotos syndrome is associated with cardiac anomalies, cloning of the breakpoints of a de novo t(5;8)(q35;q24.1) renal anomalies, seizures and/or scoliosis in B25% of translocation in a child with Sotos syndrome led to the cases and a broad variety of additional features occur discovery that Sotos syndrome is caused by haploinsuffi- less frequently. ciency of the Nuclear receptor Set Domain containing NSD1 abnormalities, such as truncating mutations, protein 1 gene, NSD1.3 Subsequently, extensive analyses of missense mutations in functional domains, partial overgrowth cases have shown that intragenic NSD1 muta- gene deletions and 5q35 microdeletions encompass- tions and 5q35 microdeletions encompassing NSD1 cause ing NSD1, are identifiable in the majority (490%) of 490% of Sotos syndrome cases.4–10 In addition, NSD1 Sotos syndrome cases. -

Insights Into the Genetic Architecture of the Human Face
bioRxiv preprint doi: https://doi.org/10.1101/2020.05.12.090555; this version posted May 14, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Insights into the genetic architecture of the human face Julie D. White1†*, Karlijne Indencleef2,3,4†*, Sahin Naqvi5,6, Ryan J. Eller7, Jasmien Roosenboom8, Myoung Keun Lee8, Jiarui Li2,3, Jaaved Mohammed5,9, Stephen Richmond10, Ellen E. Quillen11,12, Heather L. Norton13, Eleanor Feingold14, Tomek Swigut5,9, Mary L. Marazita8,14, Hilde Peeters15, Greet Hens4, John R. Shaffer8,14, Joanna Wysocka5,9, Susan Walsh7, Seth M. Weinberg8,14,16, Mark D. Shriver1, Peter Claes2,3,15,17,18* Affiliations: 1Department of Anthropology, Pennsylvania State University, State College, PA, 16802, USA 2Department of Electrical Engineering, ESAT/PSI, KU Leuven, Leuven, 3000, Belgium 3Medical Imaging Research Center, UZ Leuven, Leuven, 3000, Belgium 4Department of Otorhinolaryngology, KU Leuven, Leuven, 3000, Belgium 5Department of Chemical and Systems Biology, Stanford University School of Medicine, Stanford, CA, 94305, USA 6Department of Genetics, Stanford University School of Medicine, Stanford, CA, 94305, USA 7Department of Biology, Indiana University Purdue University Indianapolis, Indianapolis, IN, 46202, USA 8Department of Oral Biology, Center for Craniofacial and Dental Genetics, University of Pittsburgh, Pittsburgh,