Yunnan, China)
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Phaeoseptaceae, Pleosporales) from China
Mycosphere 10(1): 757–775 (2019) www.mycosphere.org ISSN 2077 7019 Article Doi 10.5943/mycosphere/10/1/17 Morphological and phylogenetic studies of Pleopunctum gen. nov. (Phaeoseptaceae, Pleosporales) from China Liu NG1,2,3,4,5, Hyde KD4,5, Bhat DJ6, Jumpathong J3 and Liu JK1*,2 1 School of Life Science and Technology, University of Electronic Science and Technology of China, Chengdu 611731, P.R. China 2 Guizhou Key Laboratory of Agricultural Biotechnology, Guizhou Academy of Agricultural Sciences, Guiyang 550006, P.R. China 3 Faculty of Agriculture, Natural Resources and Environment, Naresuan University, Phitsanulok 65000, Thailand 4 Center of Excellence in Fungal Research, Mae Fah Luang University, Chiang Rai 57100, Thailand 5 Mushroom Research Foundation, Chiang Rai 57100, Thailand 6 No. 128/1-J, Azad Housing Society, Curca, P.O., Goa Velha 403108, India Liu NG, Hyde KD, Bhat DJ, Jumpathong J, Liu JK 2019 – Morphological and phylogenetic studies of Pleopunctum gen. nov. (Phaeoseptaceae, Pleosporales) from China. Mycosphere 10(1), 757–775, Doi 10.5943/mycosphere/10/1/17 Abstract A new hyphomycete genus, Pleopunctum, is introduced to accommodate two new species, P. ellipsoideum sp. nov. (type species) and P. pseudoellipsoideum sp. nov., collected from decaying wood in Guizhou Province, China. The genus is characterized by macronematous, mononematous conidiophores, monoblastic conidiogenous cells and muriform, oval to ellipsoidal conidia often with a hyaline, elliptical to globose basal cell. Phylogenetic analyses of combined LSU, SSU, ITS and TEF1α sequence data of 55 taxa were carried out to infer their phylogenetic relationships. The new taxa formed a well-supported subclade in the family Phaeoseptaceae and basal to Lignosphaeria and Thyridaria macrostomoides. -

University of California Santa Cruz Responding to An
UNIVERSITY OF CALIFORNIA SANTA CRUZ RESPONDING TO AN EMERGENT PLANT PEST-PATHOGEN COMPLEX ACROSS SOCIAL-ECOLOGICAL SCALES A dissertation submitted in partial satisfaction of the requirements for the degree of DOCTOR OF PHILOSOPHY in ENVIRONMENTAL STUDIES with an emphasis in ECOLOGY AND EVOLUTIONARY BIOLOGY by Shannon Colleen Lynch December 2020 The Dissertation of Shannon Colleen Lynch is approved: Professor Gregory S. Gilbert, chair Professor Stacy M. Philpott Professor Andrew Szasz Professor Ingrid M. Parker Quentin Williams Acting Vice Provost and Dean of Graduate Studies Copyright © by Shannon Colleen Lynch 2020 TABLE OF CONTENTS List of Tables iv List of Figures vii Abstract x Dedication xiii Acknowledgements xiv Chapter 1 – Introduction 1 References 10 Chapter 2 – Host Evolutionary Relationships Explain 12 Tree Mortality Caused by a Generalist Pest– Pathogen Complex References 38 Chapter 3 – Microbiome Variation Across a 66 Phylogeographic Range of Tree Hosts Affected by an Emergent Pest–Pathogen Complex References 110 Chapter 4 – On Collaborative Governance: Building Consensus on 180 Priorities to Manage Invasive Species Through Collective Action References 243 iii LIST OF TABLES Chapter 2 Table I Insect vectors and corresponding fungal pathogens causing 47 Fusarium dieback on tree hosts in California, Israel, and South Africa. Table II Phylogenetic signal for each host type measured by D statistic. 48 Table SI Native range and infested distribution of tree and shrub FD- 49 ISHB host species. Chapter 3 Table I Study site attributes. 124 Table II Mean and median richness of microbiota in wood samples 128 collected from FD-ISHB host trees. Table III Fungal endophyte-Fusarium in vitro interaction outcomes. -

Molecular Systematics of the Marine Dothideomycetes
available online at www.studiesinmycology.org StudieS in Mycology 64: 155–173. 2009. doi:10.3114/sim.2009.64.09 Molecular systematics of the marine Dothideomycetes S. Suetrong1, 2, C.L. Schoch3, J.W. Spatafora4, J. Kohlmeyer5, B. Volkmann-Kohlmeyer5, J. Sakayaroj2, S. Phongpaichit1, K. Tanaka6, K. Hirayama6 and E.B.G. Jones2* 1Department of Microbiology, Faculty of Science, Prince of Songkla University, Hat Yai, Songkhla, 90112, Thailand; 2Bioresources Technology Unit, National Center for Genetic Engineering and Biotechnology (BIOTEC), 113 Thailand Science Park, Paholyothin Road, Khlong 1, Khlong Luang, Pathum Thani, 12120, Thailand; 3National Center for Biothechnology Information, National Library of Medicine, National Institutes of Health, 45 Center Drive, MSC 6510, Bethesda, Maryland 20892-6510, U.S.A.; 4Department of Botany and Plant Pathology, Oregon State University, Corvallis, Oregon, 97331, U.S.A.; 5Institute of Marine Sciences, University of North Carolina at Chapel Hill, Morehead City, North Carolina 28557, U.S.A.; 6Faculty of Agriculture & Life Sciences, Hirosaki University, Bunkyo-cho 3, Hirosaki, Aomori 036-8561, Japan *Correspondence: E.B. Gareth Jones, [email protected] Abstract: Phylogenetic analyses of four nuclear genes, namely the large and small subunits of the nuclear ribosomal RNA, transcription elongation factor 1-alpha and the second largest RNA polymerase II subunit, established that the ecological group of marine bitunicate ascomycetes has representatives in the orders Capnodiales, Hysteriales, Jahnulales, Mytilinidiales, Patellariales and Pleosporales. Most of the fungi sequenced were intertidal mangrove taxa and belong to members of 12 families in the Pleosporales: Aigialaceae, Didymellaceae, Leptosphaeriaceae, Lenthitheciaceae, Lophiostomataceae, Massarinaceae, Montagnulaceae, Morosphaeriaceae, Phaeosphaeriaceae, Pleosporaceae, Testudinaceae and Trematosphaeriaceae. Two new families are described: Aigialaceae and Morosphaeriaceae, and three new genera proposed: Halomassarina, Morosphaeria and Rimora. -

One Hundred New Species of Lichenized Fungi: a Signature of Undiscovered Global Diversity
Phytotaxa 18: 1–127 (2011) ISSN 1179-3155 (print edition) www.mapress.com/phytotaxa/ Monograph PHYTOTAXA Copyright © 2011 Magnolia Press ISSN 1179-3163 (online edition) PHYTOTAXA 18 One hundred new species of lichenized fungi: a signature of undiscovered global diversity H. THORSTEN LUMBSCH1*, TEUVO AHTI2, SUSANNE ALTERMANN3, GUILLERMO AMO DE PAZ4, ANDRÉ APTROOT5, ULF ARUP6, ALEJANDRINA BÁRCENAS PEÑA7, PAULINA A. BAWINGAN8, MICHEL N. BENATTI9, LUISA BETANCOURT10, CURTIS R. BJÖRK11, KANSRI BOONPRAGOB12, MAARTEN BRAND13, FRANK BUNGARTZ14, MARCELA E. S. CÁCERES15, MEHTMET CANDAN16, JOSÉ LUIS CHAVES17, PHILIPPE CLERC18, RALPH COMMON19, BRIAN J. COPPINS20, ANA CRESPO4, MANUELA DAL-FORNO21, PRADEEP K. DIVAKAR4, MELIZAR V. DUYA22, JOHN A. ELIX23, ARVE ELVEBAKK24, JOHNATHON D. FANKHAUSER25, EDIT FARKAS26, LIDIA ITATÍ FERRARO27, EBERHARD FISCHER28, DAVID J. GALLOWAY29, ESTER GAYA30, MIREIA GIRALT31, TREVOR GOWARD32, MARTIN GRUBE33, JOSEF HAFELLNER33, JESÚS E. HERNÁNDEZ M.34, MARÍA DE LOS ANGELES HERRERA CAMPOS7, KLAUS KALB35, INGVAR KÄRNEFELT6, GINTARAS KANTVILAS36, DOROTHEE KILLMANN28, PAUL KIRIKA37, KERRY KNUDSEN38, HARALD KOMPOSCH39, SERGEY KONDRATYUK40, JAMES D. LAWREY21, ARMIN MANGOLD41, MARCELO P. MARCELLI9, BRUCE MCCUNE42, MARIA INES MESSUTI43, ANDREA MICHLIG27, RICARDO MIRANDA GONZÁLEZ7, BIBIANA MONCADA10, ALIFERETI NAIKATINI44, MATTHEW P. NELSEN1, 45, DAG O. ØVSTEDAL46, ZDENEK PALICE47, KHWANRUAN PAPONG48, SITTIPORN PARNMEN12, SERGIO PÉREZ-ORTEGA4, CHRISTIAN PRINTZEN49, VÍCTOR J. RICO4, EIMY RIVAS PLATA1, 50, JAVIER ROBAYO51, DANIA ROSABAL52, ULRIKE RUPRECHT53, NORIS SALAZAR ALLEN54, LEOPOLDO SANCHO4, LUCIANA SANTOS DE JESUS15, TAMIRES SANTOS VIEIRA15, MATTHIAS SCHULTZ55, MARK R. D. SEAWARD56, EMMANUËL SÉRUSIAUX57, IMKE SCHMITT58, HARRIE J. M. SIPMAN59, MOHAMMAD SOHRABI 2, 60, ULRIK SØCHTING61, MAJBRIT ZEUTHEN SØGAARD61, LAURENS B. SPARRIUS62, ADRIANO SPIELMANN63, TOBY SPRIBILLE33, JUTARAT SUTJARITTURAKAN64, ACHRA THAMMATHAWORN65, ARNE THELL6, GÖRAN THOR66, HOLGER THÜS67, EINAR TIMDAL68, CAMILLE TRUONG18, ROMAN TÜRK69, LOENGRIN UMAÑA TENORIO17, DALIP K. -

Checklist of the Lichens and Allied Fungi of Kathy Stiles Freeland Bibb County Glades Preserve, Alabama, U.S.A
Opuscula Philolichenum, 18: 420–434. 2019. *pdf effectively published online 2December2019 via (http://sweetgum.nybg.org/philolichenum/) Checklist of the lichens and allied fungi of Kathy Stiles Freeland Bibb County Glades Preserve, Alabama, U.S.A. J. KEVIN ENGLAND1, CURTIS J. HANSEN2, JESSICA L. ALLEN3, SEAN Q. BEECHING4, WILLIAM R. BUCK5, VITALY CHARNY6, JOHN G. GUCCION7, RICHARD C. HARRIS8, MALCOLM HODGES9, NATALIE M. HOWE10, JAMES C. LENDEMER11, R. TROY MCMULLIN12, ERIN A. TRIPP13, DENNIS P. WATERS14 ABSTRACT. – The first checklist of lichenized, lichenicolous and lichen-allied fungi from the Kathy Stiles Freeland Bibb County Glades Preserve in Bibb County, Alabama, is presented. Collections made during the 2017 Tuckerman Workshop and additional records from herbaria and online sources are included. Two hundred and thirty-eight taxa in 115 genera are enumerated. Thirty taxa of lichenized, lichenicolous and lichen-allied fungi are newly reported for Alabama: Acarospora fuscata, A. novomexicana, Circinaria contorta, Constrictolumina cinchonae, Dermatocarpon dolomiticum, Didymocyrtis cladoniicola, Graphis anfractuosa, G. rimulosa, Hertelidea pseudobotryosa, Heterodermia pseudospeciosa, Lecania cuprea, Marchandiomyces lignicola, Minutoexcipula miniatoexcipula, Monoblastia rappii, Multiclavula mucida, Ochrolechia trochophora, Parmotrema subsumptum, Phaeographis brasiliensis, Phaeographis inusta, Piccolia nannaria, Placynthiella icmalea, Porina scabrida, Psora decipiens, Pyrenographa irregularis, Ramboldia blochiana, Thyrea confusa, Trichothelium -

Pseudodidymellaceae Fam. Nov.: Phylogenetic Affiliations Of
available online at www.studiesinmycology.org STUDIES IN MYCOLOGY 87: 187–206 (2017). Pseudodidymellaceae fam. nov.: Phylogenetic affiliations of mycopappus-like genera in Dothideomycetes A. Hashimoto1,2, M. Matsumura1,3, K. Hirayama4, R. Fujimoto1, and K. Tanaka1,3* 1Faculty of Agriculture and Life Sciences, Hirosaki University, 3 Bunkyo-cho, Hirosaki, Aomori, 036-8561, Japan; 2Research Fellow of the Japan Society for the Promotion of Science, 5-3-1 Kojimachi, Chiyoda-ku, Tokyo, 102-0083, Japan; 3The United Graduate School of Agricultural Sciences, Iwate University, 18–8 Ueda 3 chome, Morioka, 020-8550, Japan; 4Apple Experiment Station, Aomori Prefectural Agriculture and Forestry Research Centre, 24 Fukutami, Botandaira, Kuroishi, Aomori, 036-0332, Japan *Correspondence: K. Tanaka, [email protected] Abstract: The familial placement of four genera, Mycodidymella, Petrakia, Pseudodidymella, and Xenostigmina, was taxonomically revised based on morphological observations and phylogenetic analyses of nuclear rDNA SSU, LSU, tef1, and rpb2 sequences. ITS sequences were also provided as barcode markers. A total of 130 sequences were newly obtained from 28 isolates which are phylogenetically related to Melanommataceae (Pleosporales, Dothideomycetes) and its relatives. Phylo- genetic analyses and morphological observation of sexual and asexual morphs led to the conclusion that Melanommataceae should be restricted to its type genus Melanomma, which is characterised by ascomata composed of a well-developed, carbonaceous peridium, and an aposphaeria-like coelomycetous asexual morph. Although Mycodidymella, Petrakia, Pseudodidymella, and Xenostigmina are phylogenetically related to Melanommataceae, these genera are characterised by epi- phyllous, lenticular ascomata with well-developed basal stroma in their sexual morphs, and mycopappus-like propagules in their asexual morphs, which are clearly different from those of Melanomma. -

<I>Cercospora Sojina</I>
University of Tennessee, Knoxville TRACE: Tennessee Research and Creative Exchange Doctoral Dissertations Graduate School 8-2017 Genetic analysis of field populations of the plant pathogens Cercospora sojina, Corynespora cassiicola and Phytophthora colocasiae Sandesh Kumar Shrestha University of Tennessee, Knoxville, [email protected] Follow this and additional works at: https://trace.tennessee.edu/utk_graddiss Part of the Plant Pathology Commons Recommended Citation Shrestha, Sandesh Kumar, "Genetic analysis of field populations of the plant pathogens Cercospora sojina, Corynespora cassiicola and Phytophthora colocasiae. " PhD diss., University of Tennessee, 2017. https://trace.tennessee.edu/utk_graddiss/4650 This Dissertation is brought to you for free and open access by the Graduate School at TRACE: Tennessee Research and Creative Exchange. It has been accepted for inclusion in Doctoral Dissertations by an authorized administrator of TRACE: Tennessee Research and Creative Exchange. For more information, please contact [email protected]. To the Graduate Council: I am submitting herewith a dissertation written by Sandesh Kumar Shrestha entitled "Genetic analysis of field populations of the plant pathogens Cercospora sojina, Corynespora cassiicola and Phytophthora colocasiae." I have examined the final electronic copy of this dissertation for form and content and recommend that it be accepted in partial fulfillment of the equirr ements for the degree of Doctor of Philosophy, with a major in Entomology and Plant Pathology. Heather M. Young-Kelly, -

271 REFERENCES Abdullah, S.K. and Taj-Aldeen, S.J. (1989
271 REFERENCES Abdullah, S.K. and Taj-Aldeen, S.J. (1989). Extracellular enzymatic activity of aquatic and aero-aquatic conidial fungi. Hydrobiologia 174: 217–223. Abler, S.W. (2003). Ecology and taxonomy of Leptosphaerulina spp. associated with turfgrasses in the United States. M.S. Thesis. Faculty of the Virginia Polytechnic Institute and State University, Blacksburg, Virginia. Adams, D.J. (2004). Fungal cell wall chitinases and glucanases. Microbiology 150: 2029–2035. Agrios, G.N. (2005). Plant Pathology. 5th ed. Department of Plant Pathology, University of Florida. Elsevier Academic Press. Ahn, Y. (1996). Taxonomic revision of taxa originally described in Leptosphaeria from species in the Ranunculaceae, Papaveraceae and Magnoliaceae. Ph.D. Thesis. University of Illinois at Urbana-Campaign. Ainsworth, G.C. and Bisby, G.R. (1943). Dictionary of The Fungi. Wallingford, UK, CAB International. Alexopoulos, C.J., Mims, C.W. and Blackwell, M. (1996). Introductory Mycology. 4th ed. New York, John Wiley & Sons, Inc. Alias, S.A., Kuthubutheen, A.J.and Jones, E.B.G. (1995). Frequency of occurrence of fungi on wood in Malaysian mangroves. Hydrobiologia 295 : 97–106. Allen, R.B., Buchanan, P.K., Clinton, P.W. and Cone, A.J. (2000). Composition and diversity of fungi on decaying logs in a New Zealand temperate beech 272 (Nothofagus) forest. Canadian Journal of Forest Research 30: 1025–1033. Anderson, N.H. and Sedell, J.R. (1979). Detritus processing by macroinvertebrates in stream ecosystems. Annual Review of Entomology 24: 351–377. Ando, K. (1992). A Study of terrestrial aquatic Hyphomycetes. Transaction of Mycological Society of Japan 33: 415–425. Anonymous. (1995). JMP® Statistics and graphics guide. -

Redisposition of Phoma-Like Anamorphs in Pleosporales
available online at www.studiesinmycology.org STUDIES IN MYCOLOGY 75: 1–36. Redisposition of phoma-like anamorphs in Pleosporales J. de Gruyter1–3*, J.H.C. Woudenberg1, M.M. Aveskamp1, G.J.M. Verkley1, J.Z. Groenewald1, and P.W. Crous1,3,4 1CBS-KNAW Fungal Biodiversity Centre, P.O. Box 85167, 3508 AD Utrecht, The Netherlands; 2National Reference Centre, National Plant Protection Organization, P.O. Box 9102, 6700 HC Wageningen, The Netherlands; 3Wageningen University and Research Centre (WUR), Laboratory of Phytopathology, Droevendaalsesteeg 1, 6708 PB Wageningen, The Netherlands; 4Microbiology, Department of Biology, Utrecht University, Padualaan 8, 3584 CH Utrecht, The Netherlands *Correspondence: Hans de Gruyter, [email protected] Abstract: The anamorphic genus Phoma was subdivided into nine sections based on morphological characters, and included teleomorphs in Didymella, Leptosphaeria, Pleospora and Mycosphaerella, suggesting the polyphyly of the genus. Recent molecular, phylogenetic studies led to the conclusion that Phoma should be restricted to Didymellaceae. The present study focuses on the taxonomy of excluded Phoma species, currently classified inPhoma sections Plenodomus, Heterospora and Pilosa. Species of Leptosphaeria and Phoma section Plenodomus are reclassified in Plenodomus, Subplenodomus gen. nov., Leptosphaeria and Paraleptosphaeria gen. nov., based on the phylogeny determined by analysis of sequence data of the large subunit 28S nrDNA (LSU) and Internal Transcribed Spacer regions 1 & 2 and 5.8S nrDNA (ITS). Phoma heteromorphospora, type species of Phoma section Heterospora, and its allied species Phoma dimorphospora, are transferred to the genus Heterospora stat. nov. The Phoma acuta complex (teleomorph Leptosphaeria doliolum), is revised based on a multilocus sequence analysis of the LSU, ITS, small subunit 18S nrDNA (SSU), β-tubulin (TUB), and chitin synthase 1 (CHS-1) regions. -

Seifertia Shangrilaensis Sp. Nov. (Melanommataceae), a New Species from Southwest China
Phytotaxa 273 (1): 034–042 ISSN 1179-3155 (print edition) http://www.mapress.com/j/pt/ PHYTOTAXA Copyright © 2016 Magnolia Press Article ISSN 1179-3163 (online edition) http://dx.doi.org/10.11646/phytotaxa.273.1.3 Seifertia shangrilaensis sp. nov. (Melanommataceae), a new species from Southwest China JUNFU LI1,2,3,4,7, RUNGTIWA PHOOKAMSAK2,3,4,7, AUSANA MAPOOK3,4, SARANYAPHAT BOONMEE3, JARAYAMA D. BHAT5,6, KEVIN D. HYDE1,2,3,4 & SAISAMORN LUMYONG1,* 1Department of Biology, Faculty of Science, Chinag Mai University, Chiang Mai 50200, Thailand. 2World Agroforestry Centre, East and Central Asia, 132 Lanhei Road, Kunming 650201, China. 3Center of Excellence in Fungal Research, Mae Fah Luang University, Chiang Rai 57100, Thailand. 4School of Science, Mae Fah Luang University, Chiang Rai 57100, Thailand. 5Formerly, Department of Botany, Goa University, Goa-403206, India. 6128/1-J, Azad Housing Society, Curca, Goa Velha-403108, India. 7Key Laboratory of Biodiversity and Biogeography, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming 650201, China. Correspondence to*: [email protected] Abstract A new Seifertia species was isolated from hanging rachides of Rhododendron decorum in Yunnan Province, Southwest China. The new taxon was compared with the type species, S. azalea and differs in having wider conidiophores, with hya- line to subhyaline and smaller conidia, while S. azalea has olive-brown to brown, rarely branched conidiophores, and pale brown or olive-brown, very rarely septate conidia. Phylogenetic analyses of combined LSU, SSU and TEF1-α sequence data show that S. shangrilaensis forms a robust clade with S. azalea nested among the species of Melanommataceae in the order Pleosporales. -
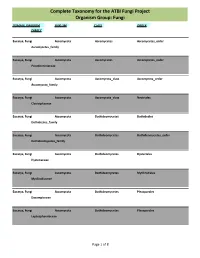
Complete Taxonomy for the ATBI Fungi Project Organism Group: Fungi
Complete Taxonomy for the ATBI Fungi Project Organism Group: Fungi DOMAIN, KINGDOM PHYLUM CLASS ORDER FAMILY Eucarya, Fungi Ascomycota Ascomycetes Ascomycetes_order Ascomycetes_family Eucarya, Fungi Ascomycota Ascomycetes Ascomycetes_order Pseudeurotiaceae Eucarya, Fungi Ascomycota Ascomycota_class Ascomycota_order Ascomycota_family Eucarya, Fungi Ascomycota Ascomycota_class Nectriales Clavicipitaceae Eucarya, Fungi Ascomycota Dothideomycetes Dothideales Dothideales_family Eucarya, Fungi Ascomycota Dothideomycetes Dothideomycetes_order Dothideomycetes_family Eucarya, Fungi Ascomycota Dothideomycetes Hysteriales Hysteriaceae Eucarya, Fungi Ascomycota Dothideomycetes Mytilinidiales Mytilinidiaceae Eucarya, Fungi Ascomycota Dothideomycetes Pleosporales Dacampiaceae Eucarya, Fungi Ascomycota Dothideomycetes Pleosporales Leptosphaeriaceae Page 1 of 8 Complete Taxonomy for the ATBI Fungi Project Organism Group: Fungi DOMAIN, KINGDOM PHYLUM CLASS ORDER FAMILY Eucarya, Fungi Ascomycota Dothideomycetes Pleosporales Lophiostomataceae Eucarya, Fungi Ascomycota Dothideomycetes Pleosporales Massarinaceae Eucarya, Fungi Ascomycota Dothideomycetes Pleosporales Melanommataceae Eucarya, Fungi Ascomycota Dothideomycetes Pleosporales Pleomassariaceae Eucarya, Fungi Ascomycota Dothideomycetes Pleosporales Pleosporaceae Eucarya, Fungi Ascomycota Dothideomycetes Pleosporales Pleosporales Eucarya, Fungi Ascomycota Dothideomycetes Pleosporales Pleosporales_family Eucarya, Fungi Ascomycota Dothideomycetes Pleosporales Sporormiaceae Eucarya, Fungi Ascomycota Dothideomycetes -

Mykologische Und Holzanatomische Untersuchungen Zur Massaria-Krankheit an Platanen
Mykologische und holzanatomische Untersuchungen zur Massaria-Krankheit an Platanen Inauguraldissertation zur Erlangung der Doktorwürde der Fakultät für Umwelt und Natürliche Ressourcen der Albert-Ludwigs-Universität Freiburg im Breisgau vorgelegt von Timo Börker Freiburg im Breisgau Februar 2019 Dekanin: Prof. Dr. D. Kleinschmit Referent: Prof. Dr. S. Fink Korreferent: Prof. Dr. F. Schwarze Zweiter Betreuer: PD Dr. B. Metzler Datum der Disputation: 06.06.2019 DANKSAGUNG Danksagung Zuerst möchte ich mich bei Herrn Prof. Dr. S. Fink für die Vergabe des Themas bedanken. Insbesondere für die Möglichkeit, die mir geboten wurde, dieses spannende Thema selbstständig mit dem nötigen Freiraum zu bearbeiten. Herrn Prof. Dr. F. Schwarze gilt mein Dank für die freundliche Übernahme des Korreferats und die damit verbundene Arbeit. Einen sehr großen Dank gehört Herrn PD Dr. B. Metzler für die Zweitbetreuung sowie Anregungen und Aufmunterungen mykologischer Fragestellungen während der gesamten Zeit. Besonders danke ich Herrn Prof. Dr. R. Kehr, der bereits während meiner Studienzeit mein Interesse für die Gehölzpathologie weckte. Seine Tipps und Anregungen waren eine große Bereicherung bei der Bearbeitung des Themas. Ganz herzlich möchte ich mich bei Herrn Dr. J. Grüner bedanken. Seine Ratschläge waren eine große Hilfe dieses Thema zu bearbeiten. Als Ansprechpartner stand er mir jederzeit zur Verfügung. Vielen, vielen Dank Jörg! Dem gesamten Team der Forstbotanik danke ich für die freundliche Unterstützung der Arbeiten. Bei Frau Dr. M. Alabed Alkader möchte ich mich für die wertvollen Tipps bei der Vorgehensweise mikrobiologischer Arbeiten bedanken. Frau S. Röske danke ich vor allem für die damit verbundenen Arbeiten der PCR, Frau N. Streit und ganz besonders Frau S.