Retinoid X Receptor Agonists Increase Bcl2a1 Expression and Decrease Apoptosis of Naive T Lymphocytes1
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

LDB3 Monoclonal Antibody (M06), Gene Alias: CYPHER, FLJ35865, KIAA01613, Clone 3C8 KIAA0613, ORACLE, PDLIM6, ZASP, Ldb3z1, Ldb3z4
LDB3 monoclonal antibody (M06), Gene Alias: CYPHER, FLJ35865, KIAA01613, clone 3C8 KIAA0613, ORACLE, PDLIM6, ZASP, ldb3z1, ldb3z4 Gene Summary: This gene encodes a PDZ Catalog Number: H00011155-M06 domain-containing protein. PDZ motifs are modular Regulatory Status: For research use only (RUO) protein-protein interaction domains consisting of 80-120 amino acid residues. PDZ domain-containing proteins Product Description: Mouse monoclonal antibody interact with each other in cytoskeletal assembly or with raised against a full-length recombinant LDB3. other proteins involved in targeting and clustering of membrane proteins. The protein encoded by this gene Clone Name: 3C8 interacts with alpha-actinin-2 through its N-terminal PDZ domain and with protein kinase C via its C-terminal LIM Immunogen: LDB3 (AAH10929, 1 a.a. ~ 283 a.a) domains. The LIM domain is a cysteine-rich motif full-length recombinant protein with GST tag. MW of the defined by 50-60 amino acids containing two GST tag alone is 26 KDa. zinc-binding modules. This protein also interacts with all three members of the myozenin family. Mutations in this Sequence: gene have been associated with myofibrillar myopathy MSYSVTLTGPGPWGFRLQGGKDFNMPLTISRITPGSK and dilated cardiomyopathy. Alternatively spliced AAQSQLSQGDLVVAIDGVNTDTMTHLEAQNKIKSASY transcript variants encoding different isoforms have been NLSLTLQKSKRPIPISTTAPPVQTPLPVIPHQKVVVNSP identified; all isoforms have N-terminal PDZ domains ANADYQERFNPSALKDSALSTHKPIEVKGLGGKATIIHA while only longer isoforms (1 and 2) have C-terminal -

Targeted Genes and Methodology Details for Neuromuscular Genetic Panels
Targeted Genes and Methodology Details for Neuromuscular Genetic Panels Reference transcripts based on build GRCh37 (hg19) interrogated by Neuromuscular Genetic Panels Next-generation sequencing (NGS) and/or Sanger sequencing is performed Motor Neuron Disease Panel to test for the presence of a mutation in these genes. Gene GenBank Accession Number Regions of homology, high GC-rich content, and repetitive sequences may ALS2 NM_020919 not provide accurate sequence. Therefore, all reported alterations detected ANG NM_001145 by NGS are confirmed by an independent reference method based on laboratory developed criteria. However, this does not rule out the possibility CHMP2B NM_014043 of a false-negative result in these regions. ERBB4 NM_005235 Sanger sequencing is used to confirm alterations detected by NGS when FIG4 NM_014845 appropriate.(Unpublished Mayo method) FUS NM_004960 HNRNPA1 NM_031157 OPTN NM_021980 PFN1 NM_005022 SETX NM_015046 SIGMAR1 NM_005866 SOD1 NM_000454 SQSTM1 NM_003900 TARDBP NM_007375 UBQLN2 NM_013444 VAPB NM_004738 VCP NM_007126 ©2018 Mayo Foundation for Medical Education and Research Page 1 of 14 MC4091-83rev1018 Muscular Dystrophy Panel Muscular Dystrophy Panel Gene GenBank Accession Number Gene GenBank Accession Number ACTA1 NM_001100 LMNA NM_170707 ANO5 NM_213599 LPIN1 NM_145693 B3GALNT2 NM_152490 MATR3 NM_199189 B4GAT1 NM_006876 MYH2 NM_017534 BAG3 NM_004281 MYH7 NM_000257 BIN1 NM_139343 MYOT NM_006790 BVES NM_007073 NEB NM_004543 CAPN3 NM_000070 PLEC NM_000445 CAV3 NM_033337 POMGNT1 NM_017739 CAVIN1 NM_012232 POMGNT2 -

Analysis of Trans Esnps Infers Regulatory Network Architecture
Analysis of trans eSNPs infers regulatory network architecture Anat Kreimer Submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy in the Graduate School of Arts and Sciences COLUMBIA UNIVERSITY 2014 © 2014 Anat Kreimer All rights reserved ABSTRACT Analysis of trans eSNPs infers regulatory network architecture Anat Kreimer eSNPs are genetic variants associated with transcript expression levels. The characteristics of such variants highlight their importance and present a unique opportunity for studying gene regulation. eSNPs affect most genes and their cell type specificity can shed light on different processes that are activated in each cell. They can identify functional variants by connecting SNPs that are implicated in disease to a molecular mechanism. Examining eSNPs that are associated with distal genes can provide insights regarding the inference of regulatory networks but also presents challenges due to the high statistical burden of multiple testing. Such association studies allow: simultaneous investigation of many gene expression phenotypes without assuming any prior knowledge and identification of unknown regulators of gene expression while uncovering directionality. This thesis will focus on such distal eSNPs to map regulatory interactions between different loci and expose the architecture of the regulatory network defined by such interactions. We develop novel computational approaches and apply them to genetics-genomics data in human. We go beyond pairwise interactions to define network motifs, including regulatory modules and bi-fan structures, showing them to be prevalent in real data and exposing distinct attributes of such arrangements. We project eSNP associations onto a protein-protein interaction network to expose topological properties of eSNPs and their targets and highlight different modes of distal regulation. -

Human and Mouse CD Marker Handbook Human and Mouse CD Marker Key Markers - Human Key Markers - Mouse
Welcome to More Choice CD Marker Handbook For more information, please visit: Human bdbiosciences.com/eu/go/humancdmarkers Mouse bdbiosciences.com/eu/go/mousecdmarkers Human and Mouse CD Marker Handbook Human and Mouse CD Marker Key Markers - Human Key Markers - Mouse CD3 CD3 CD (cluster of differentiation) molecules are cell surface markers T Cell CD4 CD4 useful for the identification and characterization of leukocytes. The CD CD8 CD8 nomenclature was developed and is maintained through the HLDA (Human Leukocyte Differentiation Antigens) workshop started in 1982. CD45R/B220 CD19 CD19 The goal is to provide standardization of monoclonal antibodies to B Cell CD20 CD22 (B cell activation marker) human antigens across laboratories. To characterize or “workshop” the antibodies, multiple laboratories carry out blind analyses of antibodies. These results independently validate antibody specificity. CD11c CD11c Dendritic Cell CD123 CD123 While the CD nomenclature has been developed for use with human antigens, it is applied to corresponding mouse antigens as well as antigens from other species. However, the mouse and other species NK Cell CD56 CD335 (NKp46) antibodies are not tested by HLDA. Human CD markers were reviewed by the HLDA. New CD markers Stem Cell/ CD34 CD34 were established at the HLDA9 meeting held in Barcelona in 2010. For Precursor hematopoetic stem cell only hematopoetic stem cell only additional information and CD markers please visit www.hcdm.org. Macrophage/ CD14 CD11b/ Mac-1 Monocyte CD33 Ly-71 (F4/80) CD66b Granulocyte CD66b Gr-1/Ly6G Ly6C CD41 CD41 CD61 (Integrin b3) CD61 Platelet CD9 CD62 CD62P (activated platelets) CD235a CD235a Erythrocyte Ter-119 CD146 MECA-32 CD106 CD146 Endothelial Cell CD31 CD62E (activated endothelial cells) Epithelial Cell CD236 CD326 (EPCAM1) For Research Use Only. -

The Role of Z-Disc Proteins in Myopathy and Cardiomyopathy
International Journal of Molecular Sciences Review The Role of Z-disc Proteins in Myopathy and Cardiomyopathy Kirsty Wadmore 1,†, Amar J. Azad 1,† and Katja Gehmlich 1,2,* 1 Institute of Cardiovascular Sciences, College of Medical and Dental Sciences, University of Birmingham, Birmingham B15 2TT, UK; [email protected] (K.W.); [email protected] (A.J.A.) 2 Division of Cardiovascular Medicine, Radcliffe Department of Medicine and British Heart Foundation Centre of Research Excellence Oxford, University of Oxford, Oxford OX3 9DU, UK * Correspondence: [email protected]; Tel.: +44-121-414-8259 † These authors contributed equally. Abstract: The Z-disc acts as a protein-rich structure to tether thin filament in the contractile units, the sarcomeres, of striated muscle cells. Proteins found in the Z-disc are integral for maintaining the architecture of the sarcomere. They also enable it to function as a (bio-mechanical) signalling hub. Numerous proteins interact in the Z-disc to facilitate force transduction and intracellular signalling in both cardiac and skeletal muscle. This review will focus on six key Z-disc proteins: α-actinin 2, filamin C, myopalladin, myotilin, telethonin and Z-disc alternatively spliced PDZ-motif (ZASP), which have all been linked to myopathies and cardiomyopathies. We will summarise pathogenic variants identified in the six genes coding for these proteins and look at their involvement in myopathy and cardiomyopathy. Listing the Minor Allele Frequency (MAF) of these variants in the Genome Aggregation Database (GnomAD) version 3.1 will help to critically re-evaluate pathogenicity based on variant frequency in normal population cohorts. -

The N-Cadherin Interactome in Primary Cardiomyocytes As Defined Using Quantitative Proximity Proteomics Yang Li1,*, Chelsea D
© 2019. Published by The Company of Biologists Ltd | Journal of Cell Science (2019) 132, jcs221606. doi:10.1242/jcs.221606 TOOLS AND RESOURCES The N-cadherin interactome in primary cardiomyocytes as defined using quantitative proximity proteomics Yang Li1,*, Chelsea D. Merkel1,*, Xuemei Zeng2, Jonathon A. Heier1, Pamela S. Cantrell2, Mai Sun2, Donna B. Stolz1, Simon C. Watkins1, Nathan A. Yates1,2,3 and Adam V. Kwiatkowski1,‡ ABSTRACT requires multiple adhesion, cytoskeletal and signaling proteins, The junctional complexes that couple cardiomyocytes must transmit and mutations in these proteins can cause cardiomyopathies (Ehler, the mechanical forces of contraction while maintaining adhesive 2018). However, the molecular composition of ICD junctional homeostasis. The adherens junction (AJ) connects the actomyosin complexes remains poorly defined. – networks of neighboring cardiomyocytes and is required for proper The core of the AJ is the cadherin catenin complex (Halbleib and heart function. Yet little is known about the molecular composition of the Nelson, 2006; Ratheesh and Yap, 2012). Classical cadherins are cardiomyocyte AJ or how it is organized to function under mechanical single-pass transmembrane proteins with an extracellular domain that load. Here, we define the architecture, dynamics and proteome of mediates calcium-dependent homotypic interactions. The adhesive the cardiomyocyte AJ. Mouse neonatal cardiomyocytes assemble properties of classical cadherins are driven by the recruitment of stable AJs along intercellular contacts with organizational and cytosolic catenin proteins to the cadherin tail, with p120-catenin β structural hallmarks similar to mature contacts. We combine (CTNND1) binding to the juxta-membrane domain and -catenin β quantitative mass spectrometry with proximity labeling to identify the (CTNNB1) binding to the distal part of the tail. -

A Shared Pathway of Exosome Biogenesis Operates at Plasma And
bioRxiv preprint doi: https://doi.org/10.1101/545228; this version posted February 11, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. A shared pathway of exosome biogenesis operates at plasma and endosome membranes Francis K. Fordjour1, George G. Daaboul2, and Stephen J. Gould1* 1Department of Biological Chemistry Johns Hopkins University Baltimore, MD USA 2Nanoview Biosciences Boston, MA USA Corresponding author: Stephen J. Gould, Ph.D. Department of Biological Chemistry Johns Hopkins University Baltimore, MD USA Email: [email protected] Tel (01) 443 847 9918 1 bioRxiv preprint doi: https://doi.org/10.1101/545228; this version posted February 11, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Summary: This study of exosome cargo protein budding reveals that cells use a common pathway for budding exosomes from plasma and endosome membranes, providing a new mechanistic explanation for exosome heterogeneity and a rational roadmap for exosome engineering. Keywords: Protein budding, tetraspanin, endosome, plasma membrane, extracellular vesicle, CD9, CD63, CD81, SPIR, interferometry Abbreviations: EV, extracellular vesicles; IB, immunoblot; IFM, immunofluorescence microscopy; IPMC, intracellular plasma membrane-connected compartment; MVB, multivesicular body; SPIR, single-particle interferometric reflectance; SPIRI, single-particle interferometric reflectance imaging 2 bioRxiv preprint doi: https://doi.org/10.1101/545228; this version posted February 11, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. -

CHD7 Represses the Retinoic Acid Synthesis Enzyme ALDH1A3 During Inner Ear Development
CHD7 represses the retinoic acid synthesis enzyme ALDH1A3 during inner ear development Hui Yao, … , Shigeki Iwase, Donna M. Martin JCI Insight. 2018;3(4):e97440. https://doi.org/10.1172/jci.insight.97440. Research Article Development Neuroscience CHD7, an ATP-dependent chromatin remodeler, is disrupted in CHARGE syndrome, an autosomal dominant disorder characterized by variably penetrant abnormalities in craniofacial, cardiac, and nervous system tissues. The inner ear is uniquely sensitive to CHD7 levels and is the most commonly affected organ in individuals with CHARGE. Interestingly, upregulation or downregulation of retinoic acid (RA) signaling during embryogenesis also leads to developmental defects similar to those in CHARGE syndrome, suggesting that CHD7 and RA may have common target genes or signaling pathways. Here, we tested three separate potential mechanisms for CHD7 and RA interaction: (a) direct binding of CHD7 with RA receptors, (b) regulation of CHD7 levels by RA, and (c) CHD7 binding and regulation of RA-related genes. We show that CHD7 directly regulates expression of Aldh1a3, the gene encoding the RA synthetic enzyme ALDH1A3 and that loss of Aldh1a3 partially rescues Chd7 mutant mouse inner ear defects. Together, these studies indicate that ALDH1A3 acts with CHD7 in a common genetic pathway to regulate inner ear development, providing insights into how CHD7 and RA regulate gene expression and morphogenesis in the developing embryo. Find the latest version: https://jci.me/97440/pdf RESEARCH ARTICLE CHD7 represses the retinoic acid synthesis enzyme ALDH1A3 during inner ear development Hui Yao,1 Sophie F. Hill,2 Jennifer M. Skidmore,1 Ethan D. Sperry,3,4 Donald L. -

Integrating Protein Copy Numbers with Interaction Networks to Quantify Stoichiometry in Mammalian Endocytosis
bioRxiv preprint doi: https://doi.org/10.1101/2020.10.29.361196; this version posted October 29, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-ND 4.0 International license. Integrating protein copy numbers with interaction networks to quantify stoichiometry in mammalian endocytosis Daisy Duan1, Meretta Hanson1, David O. Holland2, Margaret E Johnson1* 1TC Jenkins Department of Biophysics, Johns Hopkins University, 3400 N Charles St, Baltimore, MD 21218. 2NIH, Bethesda, MD, 20892. *Corresponding Author: [email protected] bioRxiv preprint doi: https://doi.org/10.1101/2020.10.29.361196; this version posted October 29, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-ND 4.0 International license. Abstract Proteins that drive processes like clathrin-mediated endocytosis (CME) are expressed at various copy numbers within a cell, from hundreds (e.g. auxilin) to millions (e.g. clathrin). Between cell types with identical genomes, copy numbers further vary significantly both in absolute and relative abundance. These variations contain essential information about each protein’s function, but how significant are these variations and how can they be quantified to infer useful functional behavior? Here, we address this by quantifying the stoichiometry of proteins involved in the CME network. We find robust trends across three cell types in proteins that are sub- vs super-stoichiometric in terms of protein function, network topology (e.g. -

Atlas Journal
Atlas of Genetics and Cytogenetics in Oncology and Haematology Home Genes Leukemias Solid Tumours Cancer-Prone Deep Insight Portal Teaching X Y 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 NA Atlas Journal Atlas Journal versus Atlas Database: the accumulation of the issues of the Journal constitutes the body of the Database/Text-Book. TABLE OF CONTENTS Volume 5, Number 2, Apr-Jun 2001 Previous Issue / Next Issue Genes COL1A1 (collagen, type I, alpha 1) (17q21.31-q22). Marie-Pierre Simon, Georges Maire, Florence Pedeutour. Atlas Genet Cytogenet Oncol Haematol 2001; 5 (2): 169-177. [Full Text] [PDF] URL : http://AtlasGeneticsOncology.org/Genes/COL1A1ID186.html NF2 (neurofibromatosis type 2) (22q12.1-12.2) - updated. James F Gusella. Atlas Genet Cytogenet Oncol Haematol 2001; 5 (2): 178-184. [Full Text] [PDF] URL : http://AtlasGeneticsOncology.org/Genes/NF2117.html PDGFB (22q12.3-q13.1). Marie-Pierre Simon, Georges Maire, Florence Pedeutour. Atlas Genet Cytogenet Oncol Haematol 2001; 5 (2): 185-194. [Full Text] [PDF] URL : http://AtlasGeneticsOncology.org/Genes/PDGFBID155.html XPA (9q22.3-9q22.3). Anne Stary, Alain Sarasin. Atlas Genet Cytogenet Oncol Haematol 2001; 5 (2): 195-204. [Full Text] [PDF] Atlas Genet Cytogenet Oncol Haematol 2001; 2 I URL : http://AtlasGeneticsOncology.org/Genes/XPAID104.html ERCC-3 (Excision repair cross-complementing rodent repair deficiency, complementation group 3) (2q21). Anne Stary, Alain Sarasin. Atlas Genet Cytogenet Oncol Haematol 2001; 5 (2): 205-214. [Full Text] [PDF] URL : http://AtlasGeneticsOncology.org/Genes/XPBID296.html XPC (3p25.1). -

Role and Regulation of the P53-Homolog P73 in the Transformation of Normal Human Fibroblasts
Role and regulation of the p53-homolog p73 in the transformation of normal human fibroblasts Dissertation zur Erlangung des naturwissenschaftlichen Doktorgrades der Bayerischen Julius-Maximilians-Universität Würzburg vorgelegt von Lars Hofmann aus Aschaffenburg Würzburg 2007 Eingereicht am Mitglieder der Promotionskommission: Vorsitzender: Prof. Dr. Dr. Martin J. Müller Gutachter: Prof. Dr. Michael P. Schön Gutachter : Prof. Dr. Georg Krohne Tag des Promotionskolloquiums: Doktorurkunde ausgehändigt am Erklärung Hiermit erkläre ich, dass ich die vorliegende Arbeit selbständig angefertigt und keine anderen als die angegebenen Hilfsmittel und Quellen verwendet habe. Diese Arbeit wurde weder in gleicher noch in ähnlicher Form in einem anderen Prüfungsverfahren vorgelegt. Ich habe früher, außer den mit dem Zulassungsgesuch urkundlichen Graden, keine weiteren akademischen Grade erworben und zu erwerben gesucht. Würzburg, Lars Hofmann Content SUMMARY ................................................................................................................ IV ZUSAMMENFASSUNG ............................................................................................. V 1. INTRODUCTION ................................................................................................. 1 1.1. Molecular basics of cancer .......................................................................................... 1 1.2. Early research on tumorigenesis ................................................................................. 3 1.3. Developing -
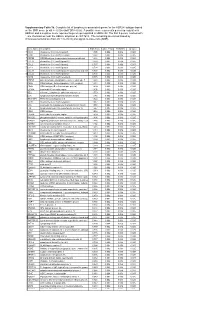
* Supplementary Table 3B. Complete List of Lymphocytic-Associated Genes
Supplementary Table 3b. Complete list of lymphocytic-associated genes for the HER2+I subtype based on the SNR score (p-val <= 0.005 and FDR<=0.05). A positive score represents genes up-regulated in HER2+I and a negative score represents genes up-regulated in HER2+NI. The first 9 genes, marked with *, are chemokines near the HER2+ amplicon at chr17q12. The remaining are sorted based by chromosomal location (from chr 1 to chr X) and signal-to-noise ratio (SNR). Gene Name Description SNR Score Feature P value FDR(BH) Q Value * CCL5 chemokine (C-C motif) ligand 5 1.395 0.002 0.036 0.025 * CCR7 chemokine (C-C motif) receptor 7 1.362 0.002 0.036 0.025 * CD79B CD79B antigen (immunoglobulin-associated beta) 1.248 0.002 0.036 0.025 * CCL13 chemokine (C-C motif) ligand 13 1.003 0.002 0.036 0.025 * CCL2 chemokine (C-C motif) ligand 2 0.737 0.004 0.056 0.037 * CCL8 chemokine (C-C motif) ligand 8 0.724 0.002 0.036 0.025 * CCL18 chemokine (C-C motif) ligand 18 (pulmonary and activ 0.703 0.002 0.036 0.025 * CCL23 chemokine (C-C motif) ligand 23 0.701 0.002 0.036 0.025 * CCR6 chemokine (C-C motif) receptor 6 0.715 0.002 0.036 0.025 PTPN7 protein tyrosine phosphatase, non-receptor type 7 1.861 0.002 0.036 0.025 CD3Z CD3Z antigen, zeta polypeptide (TiT3 complex) 1.811 0.002 0.036 0.025 CD48 CD48 antigen (B-cell membrane protein) 1.809 0.002 0.036 0.025 IL10RA interleukin 10 receptor, alpha 1.806 0.002 0.036 0.025 SELL selectin L (lymphocyte adhesion molecule 1) 1.773 0.002 0.036 0.025 LCK lymphocyte-specific protein tyrosine kinase 1.745 0.002 0.036 0.025