CD98 [19] Among Others [5][23]
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Human and Mouse CD Marker Handbook Human and Mouse CD Marker Key Markers - Human Key Markers - Mouse
Welcome to More Choice CD Marker Handbook For more information, please visit: Human bdbiosciences.com/eu/go/humancdmarkers Mouse bdbiosciences.com/eu/go/mousecdmarkers Human and Mouse CD Marker Handbook Human and Mouse CD Marker Key Markers - Human Key Markers - Mouse CD3 CD3 CD (cluster of differentiation) molecules are cell surface markers T Cell CD4 CD4 useful for the identification and characterization of leukocytes. The CD CD8 CD8 nomenclature was developed and is maintained through the HLDA (Human Leukocyte Differentiation Antigens) workshop started in 1982. CD45R/B220 CD19 CD19 The goal is to provide standardization of monoclonal antibodies to B Cell CD20 CD22 (B cell activation marker) human antigens across laboratories. To characterize or “workshop” the antibodies, multiple laboratories carry out blind analyses of antibodies. These results independently validate antibody specificity. CD11c CD11c Dendritic Cell CD123 CD123 While the CD nomenclature has been developed for use with human antigens, it is applied to corresponding mouse antigens as well as antigens from other species. However, the mouse and other species NK Cell CD56 CD335 (NKp46) antibodies are not tested by HLDA. Human CD markers were reviewed by the HLDA. New CD markers Stem Cell/ CD34 CD34 were established at the HLDA9 meeting held in Barcelona in 2010. For Precursor hematopoetic stem cell only hematopoetic stem cell only additional information and CD markers please visit www.hcdm.org. Macrophage/ CD14 CD11b/ Mac-1 Monocyte CD33 Ly-71 (F4/80) CD66b Granulocyte CD66b Gr-1/Ly6G Ly6C CD41 CD41 CD61 (Integrin b3) CD61 Platelet CD9 CD62 CD62P (activated platelets) CD235a CD235a Erythrocyte Ter-119 CD146 MECA-32 CD106 CD146 Endothelial Cell CD31 CD62E (activated endothelial cells) Epithelial Cell CD236 CD326 (EPCAM1) For Research Use Only. -

In Vitro Differentiation of Bone Marrow Mesenchymal Stem Cells Into Endometrial Epithelial Cells in Mouse: a Proteomic Analysis
Int J Clin Exp Pathol 2014;7(7):3662-3672 www.ijcep.com /ISSN:1936-2625/IJCEP0000322 Original Article In vitro differentiation of bone marrow mesenchymal stem cells into endometrial epithelial cells in mouse: a proteomic analysis Qing Cong1,2, Bin Li1,2, Yisheng Wang1,2, Wenbi Zhang1,2, Mingjun Cheng1,2, Zhiyong Wu1,2, Xiaoyan Zhang1,2, Wei Jiang1,2, Congjian Xu1,2,3,4 1Obstetrics and Gynecology Hospital of Fudan University, 2Shanghai Key Laboratory of Female Reproductive Endocrine Related Diseases, 3Department of Obstetrics and Gynecology of Shanghai Medical School, 4Institute of Biomedical Sciences, Fudan University, Shanghai, P.R. China Received March 24, 2014; Accepted June 23, 2014; Epub June 15, 2014; Published July 1, 2014 Abstract: Objective: Mouse bone marrow mesenchymal stem cells (BMSCs) have been demonstrated to differenti- ate into female endometrial epithelial cells (EECs) in vivo. Our previous studies demonstrated that BMSCs can differentiate in the direction of EECs when co-cultured with endometrial stromal cells in vitro. Here, we obtain and analyse differential proteins and their relevant pathways in the process of BMSCs differentiating into EECs by iso- baric tags for relative and absolute quantitation (iTRAQ) proteomic analysis. Methods: A 0.4-µm pore size indirect co- culture system was established with female mice endometrial stromal cells (EStCs) restricted in the upper Transwell chamber and BMSCs in the lower well plate. After indirect co-culture for several days, the BMSCs were revealed to progressively differentiate towards EECs in vitro. Then, four groups were divided according to different co-culture days with single culture groups of BMSCs as controls. -

Mechanical Forces Induce an Asthma Gene Signature in Healthy Airway Epithelial Cells Ayşe Kılıç1,10, Asher Ameli1,2,10, Jin-Ah Park3,10, Alvin T
www.nature.com/scientificreports OPEN Mechanical forces induce an asthma gene signature in healthy airway epithelial cells Ayşe Kılıç1,10, Asher Ameli1,2,10, Jin-Ah Park3,10, Alvin T. Kho4, Kelan Tantisira1, Marc Santolini 1,5, Feixiong Cheng6,7,8, Jennifer A. Mitchel3, Maureen McGill3, Michael J. O’Sullivan3, Margherita De Marzio1,3, Amitabh Sharma1, Scott H. Randell9, Jefrey M. Drazen3, Jefrey J. Fredberg3 & Scott T. Weiss1,3* Bronchospasm compresses the bronchial epithelium, and this compressive stress has been implicated in asthma pathogenesis. However, the molecular mechanisms by which this compressive stress alters pathways relevant to disease are not well understood. Using air-liquid interface cultures of primary human bronchial epithelial cells derived from non-asthmatic donors and asthmatic donors, we applied a compressive stress and then used a network approach to map resulting changes in the molecular interactome. In cells from non-asthmatic donors, compression by itself was sufcient to induce infammatory, late repair, and fbrotic pathways. Remarkably, this molecular profle of non-asthmatic cells after compression recapitulated the profle of asthmatic cells before compression. Together, these results show that even in the absence of any infammatory stimulus, mechanical compression alone is sufcient to induce an asthma-like molecular signature. Bronchial epithelial cells (BECs) form a physical barrier that protects pulmonary airways from inhaled irritants and invading pathogens1,2. Moreover, environmental stimuli such as allergens, pollutants and viruses can induce constriction of the airways3 and thereby expose the bronchial epithelium to compressive mechanical stress. In BECs, this compressive stress induces structural, biophysical, as well as molecular changes4,5, that interact with nearby mesenchyme6 to cause epithelial layer unjamming1, shedding of soluble factors, production of matrix proteins, and activation matrix modifying enzymes, which then act to coordinate infammatory and remodeling processes4,7–10. -

Exome Sequencing Identifies Mutations in the Gene TTC7A In
Developmental defects J Med Genet: first published as 10.1136/jmedgenet-2012-101483 on 19 February 2013. Downloaded from ORIGINAL ARTICLE Exome sequencing identifies mutations in the gene TTC7A in French-Canadian cases with hereditary multiple intestinal atresia Mark E Samuels,1 Jacek Majewski,2 Najmeh Alirezaie,2 Isabel Fernandez,1,3 Ferran Casals,1 Natalie Patey,1,4 Hélène Decaluwe,1,5 Isabelle Gosselin,6 Elie Haddad,1,3,5 Alan Hodgkinson,1 Youssef Idaghdour,1 Valerie Marchand,1,5 1,5 6,7 6 6 Open Access Jacques L Michaud, Marc-André Rodrigue, Sylvie Desjardins, Stéphane Dubois, Scan to access more 1,3 1,5 6,7 8 free content Francoise Le Deist, Philip Awadalla, Vincent Raymond, Bruno Maranda ▸ Additional material is ABSTRACT attempted, outcomes are poor and the condition is published online only. To view Background Congenital multiple intestinal atresia usual fatal within the first month of life. To date, please visit the journal online (http://dx.doi.org/10.1136/ (MIA) is a severe, fatal neonatal disorder, involving the no primary aetiology has been proved for the con- jmedgenet-2012-101483). occurrence of obstructions in the small and large dition. Importantly, in some cases MIA is also asso- 1 intestines ultimately leading to organ failure. Surgical ciated with either mild or severe combined Centre de Recherche du CHU fi 2–5 Ste-Justine, University of interventions are palliative but do not provide long-term immunode ciency (SCID), raising the possibility Montreal, Montreal, Quebec, survival. Severe immunodeficiency may be associated that an abnormal immune response might be the Canada with the phenotype. -

Maintenance of the Marginal Zone B Cell Compartment Specifically Requires the RNA-Binding Protein ZFP36L1
Europe PMC Funders Group Author Manuscript Nat Immunol. Author manuscript; available in PMC 2017 October 10. Published in final edited form as: Nat Immunol. 2017 June ; 18(6): 683–693. doi:10.1038/ni.3724. Europe PMC Funders Author Manuscripts Maintenance of the marginal zone B cell compartment specifically requires the RNA-binding protein ZFP36L1 Rebecca Newman1,2, Helena Ahlfors1, Alexander Saveliev1, Alison Galloway1, Daniel J Hodson3, Robert Williams1, Gurdyal S. Besra4, Charlotte N Cook5, Adam F Cunningham5, Sarah E Bell1, and Martin Turner1,* 1Laboratory of Lymphocyte Signalling and Development, The Babraham Institute, Babraham Research Campus, Cambridge, CB22 3AT, United Kingdom 2Immune Receptor Activation Laboratory, The Francis Crick Institute, 1 Midland Road, London, NW1 1AT, United Kingdom 3Department of Haematology, University of Cambridge, The Clifford Allbutt Building, Cambridge Biomedical Campus, Hills Road, Cambridge, CB2 0AH, United Kingdom 4School of Biosciences, University of Birmingham, Birmingham, B15 2TT, United Kingdom 5MRC Centre for Immune Regulation, School of Immunity and Infection, University of Birmingham, Birmingham, B15 2TT, United Kingdom Abstract Europe PMC Funders Author Manuscripts RNA binding proteins (RBP) of the ZFP36 family are best known for inhibiting the expression of cytokines through binding to AU rich elements in the 3’UTR and promoting mRNA decay. Here we show an indispensible role for ZFP36L1 as the regulator of a post-transcriptional hub that determined the identity of marginal zone (MZ) B cells by promoting their proper localization and survival. ZFP36L1 controlled a gene expression program related to signaling, cell-adhesion and locomotion, in part by limiting the expression of the transcription factors KLF2 and IRF8, which are known to enforce the follicular B cell phenotype. -
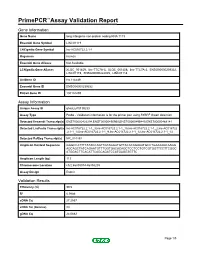
Primepcr™Assay Validation Report
PrimePCR™Assay Validation Report Gene Information Gene Name long intergenic non-protein coding RNA 1119 Ensembl Gene Symbol LINC01119 LNCipedia Gene Symbol lnc-AC016722.2.1-1 Organism Human Ensembl Gene Aliases Not Available LCNipedia Gene Aliases XLOC_001459, linc-TTC7A-3, XLOC_001458, linc-TTC7A-2, ENSG00000239332, LINC01119, ENSG00000222005, LINC01118 UniGene ID Hs.114449 Ensembl Gene ID ENSG00000239332 Entrez Gene ID 100134259 Assay Information Unique Assay ID qhsaLEP0139233 Assay Type Probe - Validation information is for the primer pair using SYBR® Green detection Detected Ensembl Transcript(s) ENST00000422294,ENST00000490950,ENST00000495449,ENST00000468141 Detected LncPedia Transcript(s) lnc-AC016722.2.1-1_3,lnc-AC016722.2.1-1_15,lnc-AC016722.2.1-1_2,lnc-AC016722 .2.1-1_14,lnc-AC016722.2.1-1_9,lnc-AC016722.2.1-1_12,lnc-AC016722.2.1-1_13 Detected RefSeq Transcript(s) NR_024452 Amplicon Context Sequence GGGCCATTTTATACCAGTTGTAGGATGTTACACAGAAATGCCTGAAAAGCAAGG AGCAGCTATCAGAATGTTTGGTGACACAGCTCCTCCTGTCGTGGTTCCTTCGGC ATGGACTTCACATTCAGCAGATCCATGAGGTGTTC Amplicon Length (bp) 113 Chromosome Location chr2:46855974-46858205 Assay Design Exonic Validation Results Efficiency (%) 99.5 R2 0.9986 cDNA Cq 27.2537 cDNA Tm (Celsius) 83 gDNA Cq 24.0682 Page 1/5 PrimePCR™Assay Validation Report Specificity (%) 100 Information to assist with data interpretation is provided at the end of this report. Page 2/5 PrimePCR™Assay Validation Report LINC01119, Human Amplification Plot Amplification of cDNA generated from 25 ng of universal reference RNA Melt Peak -

Seq2pathway Vignette
seq2pathway Vignette Bin Wang, Xinan Holly Yang, Arjun Kinstlick May 19, 2021 Contents 1 Abstract 1 2 Package Installation 2 3 runseq2pathway 2 4 Two main functions 3 4.1 seq2gene . .3 4.1.1 seq2gene flowchart . .3 4.1.2 runseq2gene inputs/parameters . .5 4.1.3 runseq2gene outputs . .8 4.2 gene2pathway . 10 4.2.1 gene2pathway flowchart . 11 4.2.2 gene2pathway test inputs/parameters . 11 4.2.3 gene2pathway test outputs . 12 5 Examples 13 5.1 ChIP-seq data analysis . 13 5.1.1 Map ChIP-seq enriched peaks to genes using runseq2gene .................... 13 5.1.2 Discover enriched GO terms using gene2pathway_test with gene scores . 15 5.1.3 Discover enriched GO terms using Fisher's Exact test without gene scores . 17 5.1.4 Add description for genes . 20 5.2 RNA-seq data analysis . 20 6 R environment session 23 1 Abstract Seq2pathway is a novel computational tool to analyze functional gene-sets (including signaling pathways) using variable next-generation sequencing data[1]. Integral to this tool are the \seq2gene" and \gene2pathway" components in series that infer a quantitative pathway-level profile for each sample. The seq2gene function assigns phenotype-associated significance of genomic regions to gene-level scores, where the significance could be p-values of SNPs or point mutations, protein-binding affinity, or transcriptional expression level. The seq2gene function has the feasibility to assign non-exon regions to a range of neighboring genes besides the nearest one, thus facilitating the study of functional non-coding elements[2]. Then the gene2pathway summarizes gene-level measurements to pathway-level scores, comparing the quantity of significance for gene members within a pathway with those outside a pathway. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

The Capacity of Long-Term in Vitro Proliferation of Acute Myeloid
The Capacity of Long-Term in Vitro Proliferation of Acute Myeloid Leukemia Cells Supported Only by Exogenous Cytokines Is Associated with a Patient Subset with Adverse Outcome Annette K. Brenner, Elise Aasebø, Maria Hernandez-Valladares, Frode Selheim, Frode Berven, Ida-Sofie Grønningsæter, Sushma Bartaula-Brevik and Øystein Bruserud Supplementary Material S2 of S31 Table S1. Detailed information about the 68 AML patients included in the study. # of blasts Viability Proliferation Cytokine Viable cells Change in ID Gender Age Etiology FAB Cytogenetics Mutations CD34 Colonies (109/L) (%) 48 h (cpm) secretion (106) 5 weeks phenotype 1 M 42 de novo 241 M2 normal Flt3 pos 31.0 3848 low 0.24 7 yes 2 M 82 MF 12.4 M2 t(9;22) wt pos 81.6 74,686 low 1.43 969 yes 3 F 49 CML/relapse 149 M2 complex n.d. pos 26.2 3472 low 0.08 n.d. no 4 M 33 de novo 62.0 M2 normal wt pos 67.5 6206 low 0.08 6.5 no 5 M 71 relapse 91.0 M4 normal NPM1 pos 63.5 21,331 low 0.17 n.d. yes 6 M 83 de novo 109 M1 n.d. wt pos 19.1 8764 low 1.65 693 no 7 F 77 MDS 26.4 M1 normal wt pos 89.4 53,799 high 3.43 2746 no 8 M 46 de novo 26.9 M1 normal NPM1 n.d. n.d. 3472 low 1.56 n.d. no 9 M 68 MF 50.8 M4 normal D835 pos 69.4 1640 low 0.08 n.d. -

(HMGB1) Deletion Leads to Small Heart and Glycolipid Metabolic
Yu et al. Cell Death Discovery (2020) 6:106 https://doi.org/10.1038/s41420-020-00340-9 Cell Death Discovery ARTICLE Open Access Cardiomyocyte-restricted high-mobility group box 1 (HMGB1) deletion leads to small heart and glycolipid metabolic disorder through GR/PGC-1α signalling Peng Yu 1, Ming Liu2,BaoliZhang3,YingYu2,EnyongSu3,ShiyaoXie3,LeiZhang3,XueYang3,HongJiang 3, Ruizhen Chen3, Yunzeng Zou3 and Junbo Ge3 Abstract Cardiac growth and remodelling are key biological processes influencing the physiological performance of the heart, and a previous study showed a critical role for intracellular HMGB1 in vitro. However, the in vivo study, which used conditional Hmgb1 ablation, did not show a significant effect on cellular or organic function. We have demonstrated the extracellular effect of HMGB1 as a pro-inflammatory molecule on cardiac remodelling. In this study, we found that HMGB1 deletion by cTnT-Cre in mouse hearts altered glucocorticoid receptor (GR) function and glycolipid metabolism, eventually leading to growth retardation, small heart and heart failure. The subcellular morphology did not show a significant change caused by HMGB1 knockout. The heart showed significant elevation of glycolysis, free fatty acid deposition and related enzyme changes. Transcriptomic analysis revealed a list of differentially expressed genes that coincide with glucocorticoid receptor function in neonatal mice and a significant increase in inflammatory genes in 1234567890():,; 1234567890():,; 1234567890():,; 1234567890():,; adult mice. Cardiac HMGB1 knockout led to a series of changes in PGC-1α, UCP3 and GyK, which were the cause of metabolic changes and further impacted cardiac function. Ckmm-Cre Hmgb1fl/fl mice did not show a specific phenotype, which was consistent with the reported negative result of cardiomyocyte-specific Hmgb1 deletion via MHC-Cre. -

A Shared Pathway of Exosome Biogenesis Operates at Plasma And
bioRxiv preprint doi: https://doi.org/10.1101/545228; this version posted February 11, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. A shared pathway of exosome biogenesis operates at plasma and endosome membranes Francis K. Fordjour1, George G. Daaboul2, and Stephen J. Gould1* 1Department of Biological Chemistry Johns Hopkins University Baltimore, MD USA 2Nanoview Biosciences Boston, MA USA Corresponding author: Stephen J. Gould, Ph.D. Department of Biological Chemistry Johns Hopkins University Baltimore, MD USA Email: [email protected] Tel (01) 443 847 9918 1 bioRxiv preprint doi: https://doi.org/10.1101/545228; this version posted February 11, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Summary: This study of exosome cargo protein budding reveals that cells use a common pathway for budding exosomes from plasma and endosome membranes, providing a new mechanistic explanation for exosome heterogeneity and a rational roadmap for exosome engineering. Keywords: Protein budding, tetraspanin, endosome, plasma membrane, extracellular vesicle, CD9, CD63, CD81, SPIR, interferometry Abbreviations: EV, extracellular vesicles; IB, immunoblot; IFM, immunofluorescence microscopy; IPMC, intracellular plasma membrane-connected compartment; MVB, multivesicular body; SPIR, single-particle interferometric reflectance; SPIRI, single-particle interferometric reflectance imaging 2 bioRxiv preprint doi: https://doi.org/10.1101/545228; this version posted February 11, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. -

Actin Binding LIM 1 (Ablim1) Negatively Controls Osteoclastogenesis by Regulating Cell Migration and Fusion
Received: 14 September 2017 | Accepted: 16 March 2018 DOI: 10.1002/jcp.26605 ORIGINAL RESEARCH ARTICLE Actin binding LIM 1 (abLIM1) negatively controls osteoclastogenesis by regulating cell migration and fusion Haruna Narahara1,2 | Eiko Sakai1 | Yu Yamaguchi1 | Shun Narahara1 | Mayumi Iwatake1,* | Kuniaki Okamoto1,† | Noriaki Yoshida2 | Takayuki Tsukuba1 1 Department of Dental Pharmacology, Graduate School of Biomedical Sciences, Actin binding LIM 1 (abLIM1) is a cytoskeletal actin-binding protein that has been Nagasaki University, Nagasaki, Japan implicated in interactions between actin filaments and cytoplasmic targets. Previous 2 Department of Orthodontics and Dentofacial Orthopedics, Graduate School of Biomedical biochemical and cytochemical studies have shown that abLIM1 interacts and co- Sciences, Nagasaki University, Nagasaki, Japan localizes with F-actin in the retina and muscle. However, whether abLIM1 regulates Correspondence osteoclast differentiation has not yet been elucidated. In this study, we examined the Takayuki Tsukuba, Department of Dental role of abLIM1 in osteoclast differentiation and function. We found that abLIM1 Pharmacology, Graduate School of Biomedical Sciences, Nagasaki University, Nagasaki 852- expression was upregulated during receptor activator of nuclear factor kappa-B ligand 8588, Japan. (RANKL)-induced osteoclast differentiation, and that a novel transcript of abLIM1 was Email: [email protected] exclusively expressed in osteoclasts. Overexpression of abLIM1 in the murine Funding information monocytic cell line, RAW-D suppressed osteoclast differentiation and decreased JSPS KAKENHI, Grant numbers: 15H05298, 16K15790, 17H04379 expression of several osteoclast-marker genes. By contrast, small interfering RNA-induced knockdown of abLIM1 enhanced the formation of multinucleated osteoclasts and markedly increased the expression of the osteoclast-marker genes. Mechanistically, abLIM1 regulated the localization of tubulin, migration, and fusion in osteoclasts.