Hepatitis E Outbreak in the Central Part of Italy Sustained by Multiple HEV Genotype 3 Strains, June–December 2019
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
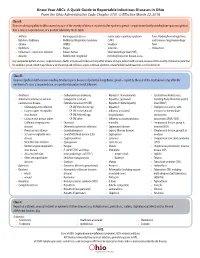
The Know Your Abcs: a Quick Guide to Reportable Infectious Diseases
Know Your ABCs: A Quick Guide to Reportable Infectious Diseases in Ohio From the Ohio Administrative Code Chapter 3701-3; Effective March 22, 2018 Class A: Diseases of major public health concern because of the severity of disease or potential for epidemic spread – report immediately via telephone upon recognition that a case, a suspected case, or a positive laboratory result exists. • Anthrax • Meningococcal disease • Severe acute respiratory syndrome fever, Marburg hemorrhagic fever, • Botulism, foodborne • Middle East Respiratory Syndrome (SARS) and Crimean-Congo hemorrhagic • Cholera (MERS) • Smallpox fever • Diphtheria • Plague • Tularemia • Yellow fever • Influenza A – novel virus infection • Rabies, human • Viral hemorrhagic fever (VHF), • Measles • Rubella (not congenital) including Ebola virus disease, Lassa Any unexpected pattern of cases, suspected cases, deaths or increased incidence of any other disease of major public health concern, because of the severity of disease or potential for epidemic spread, which may indicate a newly recognized infectious agent, outbreak, epidemic, related public health hazard or act of bioterrorism. Class B: Disease of public health concern needing timely response because of potential for epidemic spread – report by the end of the next business day after the existence of a case, a suspected case, or a positive laboratory result is known. • Amebiasis • Carbapenemase-producing • Hepatitis C (non-perinatal) • Spotted Fever Rickettsiosis, • Arboviral neuroinvasive and non- carbapenem-resistant • Hepatitis C (perinatal) including Rocky Mountain spotted neuroinvasive disease: Enterobacteriaceae (CP-CRE) • Hepatitis D (delta hepatitis) fever (RMSF) • Chikungunya virus infection • CP-CRE Enterobacter spp. • Hepatitis E • Staphylococcus aureus, with • Eastern equine encephalitis • CP-CRE Escherichia coli • Influenza-associated resistance or intermediate virus disease • CP-CRE Klebsiella spp. -

Reportable Diseases and Conditions
KINGS COUNTY DEPARTMENT of PUBLIC HEALTH 330 CAMPUS DRIVE, HANFORD, CA 93230 REPORTABLE DISEASES AND CONDITIONS Title 17, California Code of Regulations, §2500, requires that known or suspected cases of any of the diseases or conditions listed below are to be reported to the local health jurisdiction within the specified time frame: REPORT IMMEDIATELY BY PHONE During Business Hours: (559) 852-2579 After Hours: (559) 852-2720 for Immediate Reportable Disease and Conditions Anthrax Escherichia coli: Shiga Toxin producing (STEC), Rabies (Specify Human or Animal) Botulism (Specify Infant, Foodborne, Wound, Other) including E. coli O157:H7 Scrombroid Fish Poisoning Brucellosis, Human Flavivirus Infection of Undetermined Species Shiga Toxin (Detected in Feces) Cholera Foodborne Disease (2 or More Cases) Smallpox (Variola) Ciguatera Fish Poisoning Hemolytic Uremic Syndrome Tularemia, human Dengue Virus Infection Influenza, Novel Strains, Human Viral Hemorrhagic Fever (Crimean-Congo, Ebola, Diphtheria Measles (Rubeola) Lassa, and Marburg Viruses) Domonic Acid Poisoning (Amnesic Shellfish Meningococcal Infections Yellow Fever Poisoning) Novel Virus Infection with Pandemic Potential Zika Virus Infection Paralytic Shellfish Poisoning Plague (Specify Human or Animal) Immediately report the occurrence of any unusual disease OR outbreaks of any disease. REPORT BY PHONE, FAX, MAIL WITHIN ONE (1) WORKING DAY Phone: (559) 852-2579 Fax: (559) 589-0482 Mail: 330 Campus Drive, Hanford 93230 Conditions may also be reported electronically via the California -
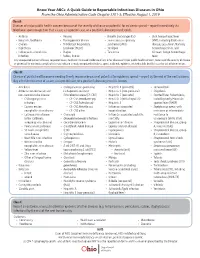
Know Your Abcs: a Quick Guide to Reportable Infectious Diseases in Ohio
Know Your ABCs: A Quick Guide to Reportable Infectious Diseases in Ohio From the Ohio Administrative Code Chapter 3701-3; Effective August 1, 2019 Class A: Diseases of major public health concern because of the severity of disease or potential for epidemic spread – report immediately via telephone upon recognition that a case, a suspected case, or a positive laboratory result exists. • Anthrax • Measles • Rubella (not congenital) • Viral hemorrhagic fever • Botulism, foodborne • Meningococcal disease • Severe acute respiratory (VHF), including Ebola virus • Cholera • Middle East Respiratory syndrome (SARS) disease, Lassa fever, Marburg • Diphtheria Syndrome (MERS) • Smallpox hemorrhagic fever, and • Influenza A – novel virus • Plague • Tularemia Crimean-Congo hemorrhagic infection • Rabies, human fever Any unexpected pattern of cases, suspected cases, deaths or increased incidence of any other disease of major public health concern, because of the severity of disease or potential for epidemic spread, which may indicate a newly recognized infectious agent, outbreak, epidemic, related public health hazard or act of bioterrorism. Class B: Disease of public health concern needing timely response because of potential for epidemic spread – report by the end of the next business day after the existence of a case, a suspected case, or a positive laboratory result is known. • Amebiasis • Carbapenemase-producing • Hepatitis B (perinatal) • Salmonellosis • Arboviral neuroinvasive and carbapenem-resistant • Hepatitis C (non-perinatal) • Shigellosis -

Hepatitis E Virus Genotype 3 in Shellfish, United Kingdom
LETTERS term to dramatically reduce the high 3. Biswas PK, Christensen JP, Ahmed SS, biologically relevant sentinels and incidence of HPAI in Bangladesh. We Barua H, Das A, Rahman MH. at al. Avian can indicate potential pathogens that infl uenza outbreaks in chickens, Bangla- have progressively and dramatically desh. Emerg Infect Dis. 2008;14:1909–12. are contaminating the environment. increased the scope and benefi ts of our http://dx.doi.org/10.3201/eid1412.071567 It is essential to ensure that this pilot PVC implementation program, 4. Biswas PK, Christensen JP, Ahmed SS, sustainable resource of coastal areas, but additional work is needed. To Das A, Rahman MH, Barua H, et al. where mussels and oysters are farmed Risk for infection with highly pathogenic help spread PVC approaches through- avian infl uenza virus (H5N1) in back- or collected wild, is not subjected to out the country, community leaders, yard chickens, Bangladesh. Emerg In- environmental contamination that imams of local mosques, and school fect Dis. 2009;15:1931–6. http://dx.doi. could lead to public health risks. teachers can be trained to implement org/10.3201/eid1512.090643 Risk management for bivalve 5. Biswas PK, Rahman MH, Das A, Ahmed awareness programs on safe practices SS, Giasuddin M, Christensen JP. Risk mollusks, aimed at control of fecal for raising poultry and regular clean- for highly pathogenic avian infl uenza pollution, relies heavily on the use ing and disinfection of live bird mar- H5N1 virus infection in chickens in of Escherichia coli as an indicator kets. The strengthening of biosecurity small-scale commercial farms, in a high- of fecal (sewage) contamination risk area, Bangladesh, 2008. -

Emerging Water-Borne Pathogens
Appl Microbiol Biotechnol (2003) 61:424–428 DOI 10.1007/s00253-003-1302-y MINI-REVIEW S. Sharma · P. Sachdeva · J. S. Virdi Emerging water-borne pathogens Received: 1 October 2002 / Revised: 27 February 2003 / Accepted: 28 February 2003 / Published online: 9 April 2003 Springer-Verlag 2003 Abstract Emerging water-borne pathogens constitute a number of reviews and other publications available on the major health hazard in both developed and developing subject discuss emerging and re-emerging pathogens in nations. A new dimension to the global epidemiology of general, including water-borne, without addressing the cholera—an ancient scourge—was provided by the issues specifically pertinent to emerging water-borne emergence of Vibrio cholerae O139. Also, water-borne pathogens. This review critically examines which organ- enterohaemorrhagic Escherichia coli (E. coli O157:H7), isms really qualify as emerging water-borne pathogens, although regarded as a problem of the industrialized west, the possible reasons underlying their emergence and has recently caused outbreaks in Africa. Outbreaks of specific measures to face the challenge posed by them. chlorine-resistant Cryptosporidium have motivated water Due to the paucity of data, it may be difficult to decide authorities to reassess the adequacy of current water- which of all water-borne pathogens are emerging. Nev- quality regulations. Of late, a host of other organisms, ertheless, there are some clear-cut candidates. such as hepatitis viruses (including hepatitis E virus), No organism other than Vibrio cholerae could serve as Campylobacter jejuni, microsporidia, cyclospora, Yersin- a better example of an emerging water-borne pathogen. ia enterocolitica, calciviruses and environmental bacteria Cholera is an ancient scourge and to date seven like Mycobacterium spp, aeromonads, Legionella pneu- pandemics have been recorded. -

Disease Reporting-Clinician
Infections, diseases and conditions reportable by clinicians: 2015 Report immediately • Anthrax (Bacillus anthracis) • Botulism (Clostridium botulinum) • Cholera (Vibrio cholerae O1, O139, or toxigenic) • Diphtheria (Corynebacterium diphtheriae) • Hemorrhagic fever caused by viruses of the filovirus (e.g., Ebola, Marburg) or arenavirus (e.g., Lassa, Machupo) families • Influenza (novel)1 (From footnote: Influenza A virus that cannot be subtyped by commercially distributed assays) • Marine intoxication (intoxication caused by marine microorganisms or their byproducts (e.g., paralytic shellfish poisoning, domoic acid intoxication, ciguatera, scombroid) • Measles (rubeola) • Plague (Yersinia pestis) • Poliomyelitis • Rabies (human) • Rubella • SARS (Severe Acute Respiratory Syndrome or SARS-coronavirus) • Smallpox (variola) • Tularemia (Francisella tularensis) • Yellow fever • Outbreaks and uncommon illnesses (any known or suspected common-source outbreak; any uncommon illness of potential public health significance) Report within 24 hours (including weekends and holidays) • Haemophilus influenzae (any isolation or identification from a normally sterile specimen type) • Neisseria meningitidis • Pesticide poisoning Public Health Division | Infections, diseases and conditions reportable by clinicians 128 Report within one working day • Amebic infections (central nervous system only) (for example infection by Naegleria or Balamuthia spp. • Animal bites (of humans) • Arthropod vector-borne disease (babesiosis, California encephalitis, Colorado -

Infant Botulism ESCMID Online Lecture Library
Emerging issues in food-borne infection ‘We never look for it because we never find it’ – emerging pathogens we need to know about ©John by Threlfall author ESCMID Online UKLecture Library ECCMID April 2013 ‘We never look for it because we never find it’ – emerging pathogens we need to know about ‘There are known knowns; there are things we know we know. We also know there are known unknowns; that is to say, we know there are some things we do not know’. ‘But there are also unknown unknowns – the ones we don’t know we don’t know’.© by author ESCMID Online Lecture Library Donald Rumsfeld, US Defence Secretary, 2002 ‘We never look for it because we never find it’ - emerging pathogens we need to know about Questions (1) 1. What is the trigger for recognising the emergence of a ‘new’ pathogen? Possible answers: Increase in incidence above ‘expected’ norm. Outbreak of ‘new’ strain. Increase in mortality in vulnerable population groups. 2. Which ‘emerging pathogen’© by hasauthor been responsible for major outbreak(s) of food-poisoning in Europe over the last five years, involving numerous fatalities? ESCMID Online Lecture Library Possible answers: Norovirus Cryptosporidium Hepatitis E (HEV) Escherichia coli O104:H4 ‘We never look for it because we never find it’ – emerging pathogens we need to know about Disease / Organism • What are the disease symptoms? • What is the causative organism? • How rapidly is this isolated? • How common is the organism? – individuals, communities, environment Source / Reservoir © by author • Is it a food contamination issue? • ESCMIDIf so, what is the food? Online Lecture Library • What is the food animal reservoir (if any?) ‘We never look for it because we never find it’ - emerging pathogens we need to know about Questions (2) 1. -

Epidemiologic Notes and Reports Hepatitis E Among U.S. Travelers
January 15, 1993 / Vol. 42 / No. 1 CENTERS FOR DISEASE CONTROL AND PREVENTION 1 Hepatitis E Among U.S. Travelers, 1989–1992 4 Surveillance of Deaths Attributed to a Nor’easter — December 1992 5 Respiratory Syncytial Virus Outbreak Activity — United States, 1992 7 Condom Use and Sexual Identity Among Men Who Have Sex With Men — Dallas, 1991 14 Surveillance of the Health Status of Bhutanese Refugees — Nepal, 1992 17 Notice to Readers Epidemiologic Notes and Reports Hepatitis E Among U.S. Travelers, 1989–1992 HepatitisOutbreaks E of— hepatitisContinued E (i.e., enterically transmitted non-A, non-B hepatitis) have occurred in some parts of the world and have generally been related to contaminated water supplies. Until recently, when research-based serologic tests (1,2 ) were devel- oped to test for antibody to hepatitis E virus (anti-HEV), no serologic test was available to identify HEV infection, and diagnosis depended on a history of exposure in an ap- propriate epidemiologic setting and the exclusion of other causes of viral hepatitis. During 1989–1992, acute HEV infection was documented among six persons in the United States who had returned from international travel. This report summarizes CDC’s serologic documentation of acute HEV infection—presumed to have been ac- quired during international travel—in four of these persons. Patient 1 On February 23, 1991, a woman from Denver traveled to Rosarito Beach, Mexico, for 1 day (3 ). On March 17, she developed headache and nausea, and on March 23, became jaundiced. A serum specimen obtained on March 23 demonstrated a serum aspartate aminotransferase (AST) level of 2100 U/L (normal: 0–35 U/L), an alkaline phosphatase level of 516 U/L (normal: 110–295 U/L), and a total bilirubin level of 7.5 mg/dL (normal: 0–1 mg/dL). -

Hepatitis a and Hepatitis E Luis S
Hepatitis A and Hepatitis E Luis S. Marsano, MD Professor of Medicine Division of Gastroenterology, Hepatology & Nutrition University of Louisville and Louisville VAMC 2011 Viruses that cause Hepatitis in Humans • Hepatitis A • Lassa virus • Hepatitis B • Rift Valley virus • Hepatitis C • Marburg virus • Hepatitis D • Ebola virus • Hepatitis E • Measles virus • Coxsackie • Human adenovirus • Echovirus • CMV • Yellow fever • EBV • Rubella • HSV • Junin virus (Argentina) • Varicella‐Zoster • Machupo virus (Bolivia) Hepatitis A Hepatitis A • Hepatovirus from Picornavirus family. • 27‐32 nm, linear, (+) sense, simple stranded RNA. • Reservoir: human only • Acquisition: fecal‐oral; concentrated in oysters. Contaminated water or food, men having sex with men, blood during short viremic phase. • Non‐cytopatic; immune mediated injury by lymphocytes Groups at Risk for HAV • Healthy persons who travel to endemic areas, • Workers in occupations with high likelihood of exposure, • Family members of infected patients, • Persons who adopt infants or children from endemic areas, • Men who have sex with men, • Persons who have tested positive for human immunodeficiency virus, • Persons with chronic liver disease, • Persons with clotting factor disorders, • Users of injection and noninjection illicit drugs. Hepatitis A Sero‐Prevalence • Poor sanitation countries: – near 100 % by age 5. • Good sanitation countries (USA) : – 10 % by age 14; – 37 % in adults. Hepatitis A Clinical Features • Incubation: 2‐4 (up to 6) weeks • Children < 2 year: 80% anicteric & asymptomatic • Children > 2 y and adults: 80% icteric & symptomatic • Symptoms: Fatigue (90%), anorexia (85%), jaundice (80%), nausea (75%), low fever (65%), headache, abdominal pain and myalgias. • Duration: usually < 8 weeks Hepatitis A Atypical Manifestations • Relapsing hepatitis: less than 10%; 2 or more bouts of elevated enzymes. -

Quantitative Risk Profile for Viruses in Foods
Quantitative risk profile for viruses in foods RIVM report 330371008/2013 M. Bouwknegt et al. National Institute for Public Health and the Environment P.O. Box 1 | 3720 BA Bilthoven www.rivm.com Quantitative risk profile for viruses in foods RIVM Report 330371008/2013 RIVM Report 330371008 Colophon © RIVM 2013 Parts of this publication may be reproduced, provided acknowledgement is given to the 'National Institute for Public Health and the Environment', along with the title and year of publication. M. Bouwknegt, LZO, RIVM/CIb K. Verhaelen, LZO, RIVM/CIb A.M. de Roda Husman, LZO, RIVM/CIb S.A. Rutjes, LZO, RIVM/CIb Contact: Saskia Rutjes Laboratory for Zoonoses and Environmental Microbiology [email protected] This investigation has been performed by order and for the account of Food- and Consumer Product Safety Authority, within the framework of V/330371/01/VV: viruses in food. Page 2 of 64 RIVM Report 330371008 Abstract Quantitative risk profile for viruses in food Viruses, similar to bacteria, can pose a risk to human health when present in food, but comparatively little is known about them in this context. In a study aimed at the health risks posed by viruses in food products, the RIVM has inventoried both current knowledge and pertinent information that is lacking. The inventory, which is presented in this report as a so-called risk profile, focuses on three viruses that can be transmitted to humans through food consumption. These are the hepatitis A viruses in shellfish, noroviruses in fresh fruits and vegetables and hepatitis E viruses in pork. The study was commissioned by the Dutch Food and Consumer Product Safety Authority. -

EPI GRAM September, 2015
EPI GRAM September, 2015 A Monthly Publication of the Stark Public Health Infrastructure Coalition EPI Gram is a monthly publication of the Stark County Public Health Infrastructure Coalition. It contains a summary of provisional communicable disease reports and other key public health indicators, with summary tables for Stark County, Ohio. Some reportable conditions may be under investigation and, at any given time, data may fluctuate from month to month for a specific category. Monthly Highlight: Creutzfeldt-Jakob disease (CJD) is a rapidly progressive, always fatal neurodegenerative disorder believed to be caused by abnormal versions of a type of protein called a prion. Normally these proteins are harmless; however, when they're misshapen, they become infectious and can harm normal biological processes. CJD occurs worldwide, and the estimated annual incidence in many countries, including the United States, has been reported to be about 1 to 2 cases per million population. Although a case definition has not been established by the CDC, reports should be based upon the clinical signs and symptoms (after ruling out other forms of dementia, infections, toxic and metabolic encephalopathies and tumors) and laboratory criteria identifying the 14-3-3 protein. Symptoms include confusion, poor concentration, lethargy, progressive dementia, intermittent unsteadiness and variable ataxia. CJD is a Class B reportable disease in Ohio. A 10-year history of cases shows an average of 1 case/year among the four health jurisidictions in Stark County. Thirty-six percent of the cases were confirmed based on clinical and laboratory criteria. The remaing 64% were classified as either probable or suspect cases. -

Hepatitis E Division of Disease Control What Do I Need to Know?
Division of Disease Control What Do I Need To Know? Hepatitis E What is hepatitis E? Hepatitis E is an infection of the liver caused by the hepatitis E virus. Hepatitis E is not common in North Dakota or in the United States. Who is at risk for hepatitis E? Anyone can get hepatitis E, but those at greater risk include travelers to developing countries, particularly parts of Mexico and countries in South Asia and North Africa. Most outbreaks in developing countries have been associated with contaminated drinking water. Hepatitis E is more common among people ages 15 to 40. Rare cases have occurred in the United States among people with no history of travel to countries where hepatitis E is common. What are the symptoms of hepatitis E? People with hepatitis E may not have symptoms. If symptoms do occur, they are similar to those of other types of viral hepatitis and may include tiredness, loss of appetite, nausea, abdominal discomfort, vomiting, dark urine or jaundice (i.e., yellowing of skin or whites of eyes). The severity of symptoms increases with age. Hepatitis E is more severe among pregnant women, especially in the third trimester. How soon do symptoms appear? Symptoms may appear 15 days to 64 days after exposure, but symptoms usually appear within 26 to 42 days. How is hepatitis E spread? Hepatitis E virus is found in the stools (feces) of infected people. The virus is spread by eating or drinking contaminated food or water. Transmission from person to person occurs less commonly than with hepatitis A virus.