Quantitative Risk Profile for Viruses in Foods
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Noroviruses: Q&A
University of California, Berkeley 2222 Bancroft Way Berkeley, CA 94720 Appointments 510/642-2000 Online Appointment www.uhs.berkeley.edu Noroviruses: Q&A What are noroviruses? Noroviruses are a group of viruses that cause the “stomach flu” or gastroenteritis (GAS-tro-enter-I-tis) in people. The term “norovirus” was recently approved as the official name for this group of viruses. Several other names have been used for noroviruses, including: • Norwalk-like viruses (NLVs) • caliciviruses (because they belong to the virus family Caliciviridae) • small round structured viruses. Viruses are very different from bacteria and parasites, some of which can cause illnesses similar to norovirus infection. Viruses are much smaller, are not affected by treatment with antibiotics, and cannot grow outside of a person’s body. What are the symptoms of illness caused by noroviruses? The symptoms of norovirus illness usually include nausea, vomiting, diarrhea, and some stomach cramping. Sometimes people additionally have a low-grade fever, chills, headache, muscle aches and a general sense of tiredness. The illness often begins suddenly, and the infected person may feel very sick. The illness is usually brief, with symptoms lasting only about one or two days. In general, children experience more vomiting than adults. Most people with norovirus illness have both of these symptoms. What is the name of the illness caused by noroviruses? Illness caused by norovirus infection has several names, including: • stomach flu – this “stomach flu” is not related to the flu (or influenza), which is a respiratory illness caused by influenza virus • viral gastroenteritis – the most common name for illness caused by norovirus. -
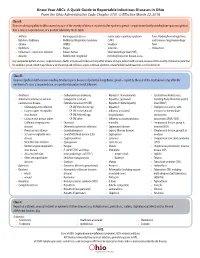
The Know Your Abcs: a Quick Guide to Reportable Infectious Diseases
Know Your ABCs: A Quick Guide to Reportable Infectious Diseases in Ohio From the Ohio Administrative Code Chapter 3701-3; Effective March 22, 2018 Class A: Diseases of major public health concern because of the severity of disease or potential for epidemic spread – report immediately via telephone upon recognition that a case, a suspected case, or a positive laboratory result exists. • Anthrax • Meningococcal disease • Severe acute respiratory syndrome fever, Marburg hemorrhagic fever, • Botulism, foodborne • Middle East Respiratory Syndrome (SARS) and Crimean-Congo hemorrhagic • Cholera (MERS) • Smallpox fever • Diphtheria • Plague • Tularemia • Yellow fever • Influenza A – novel virus infection • Rabies, human • Viral hemorrhagic fever (VHF), • Measles • Rubella (not congenital) including Ebola virus disease, Lassa Any unexpected pattern of cases, suspected cases, deaths or increased incidence of any other disease of major public health concern, because of the severity of disease or potential for epidemic spread, which may indicate a newly recognized infectious agent, outbreak, epidemic, related public health hazard or act of bioterrorism. Class B: Disease of public health concern needing timely response because of potential for epidemic spread – report by the end of the next business day after the existence of a case, a suspected case, or a positive laboratory result is known. • Amebiasis • Carbapenemase-producing • Hepatitis C (non-perinatal) • Spotted Fever Rickettsiosis, • Arboviral neuroinvasive and non- carbapenem-resistant • Hepatitis C (perinatal) including Rocky Mountain spotted neuroinvasive disease: Enterobacteriaceae (CP-CRE) • Hepatitis D (delta hepatitis) fever (RMSF) • Chikungunya virus infection • CP-CRE Enterobacter spp. • Hepatitis E • Staphylococcus aureus, with • Eastern equine encephalitis • CP-CRE Escherichia coli • Influenza-associated resistance or intermediate virus disease • CP-CRE Klebsiella spp. -

Reportable Diseases and Conditions
KINGS COUNTY DEPARTMENT of PUBLIC HEALTH 330 CAMPUS DRIVE, HANFORD, CA 93230 REPORTABLE DISEASES AND CONDITIONS Title 17, California Code of Regulations, §2500, requires that known or suspected cases of any of the diseases or conditions listed below are to be reported to the local health jurisdiction within the specified time frame: REPORT IMMEDIATELY BY PHONE During Business Hours: (559) 852-2579 After Hours: (559) 852-2720 for Immediate Reportable Disease and Conditions Anthrax Escherichia coli: Shiga Toxin producing (STEC), Rabies (Specify Human or Animal) Botulism (Specify Infant, Foodborne, Wound, Other) including E. coli O157:H7 Scrombroid Fish Poisoning Brucellosis, Human Flavivirus Infection of Undetermined Species Shiga Toxin (Detected in Feces) Cholera Foodborne Disease (2 or More Cases) Smallpox (Variola) Ciguatera Fish Poisoning Hemolytic Uremic Syndrome Tularemia, human Dengue Virus Infection Influenza, Novel Strains, Human Viral Hemorrhagic Fever (Crimean-Congo, Ebola, Diphtheria Measles (Rubeola) Lassa, and Marburg Viruses) Domonic Acid Poisoning (Amnesic Shellfish Meningococcal Infections Yellow Fever Poisoning) Novel Virus Infection with Pandemic Potential Zika Virus Infection Paralytic Shellfish Poisoning Plague (Specify Human or Animal) Immediately report the occurrence of any unusual disease OR outbreaks of any disease. REPORT BY PHONE, FAX, MAIL WITHIN ONE (1) WORKING DAY Phone: (559) 852-2579 Fax: (559) 589-0482 Mail: 330 Campus Drive, Hanford 93230 Conditions may also be reported electronically via the California -

Fact Sheet Norovirus
New Hampshire Department of Health and Human Services Fact Sheet Division of Public Health Services Norovirus What is norovirus? How is norovirus infection diagnosed? Noroviruses are a group of viruses that cause Laboratory diagnosis is difficult but there are the “stomach flu,” or gastrointestinal tests that can be performed in the New (stomach and digestive) illness. Norovirus Hampshire Public Health Lab in situations infection occurs occasionally in only one or a where there are multiple cases. Diagnosis is few people or it can be responsible for large often based on the combination of symptoms outbreaks, such as in long-term care facilities. and the short time of the illness. Who gets norovirus? What is the treatment for norovirus Norovirus infects people of all ages infection? worldwide. It may, however, be more No specific treatment is available. People who common in adults and older children. become dehydrated might need to be rehydrated by taking liquids by mouth. How does someone get norovirus? Occasionally patients may need to be Norovirus is spread from person to person via hospitalized to receive intravenous fluids. feces, but some evidence suggests that the virus is spread through the air during How can norovirus be prevented? vomiting. Good hand washing is the most While there is no vaccine for norovirus, there important way to prevent the transmission of are precautions people should take: norovirus. Outbreaks have been linked to sick • Wash hands with soap and warm water food handlers, ill health care workers, cases in after using the bathroom and after facilities such as nursing homes spreading to changing diapers other residents, contaminated shellfish, and • Wash hands with soap and warm water water contaminated with sewage. -
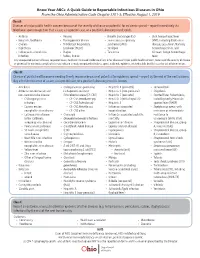
Know Your Abcs: a Quick Guide to Reportable Infectious Diseases in Ohio
Know Your ABCs: A Quick Guide to Reportable Infectious Diseases in Ohio From the Ohio Administrative Code Chapter 3701-3; Effective August 1, 2019 Class A: Diseases of major public health concern because of the severity of disease or potential for epidemic spread – report immediately via telephone upon recognition that a case, a suspected case, or a positive laboratory result exists. • Anthrax • Measles • Rubella (not congenital) • Viral hemorrhagic fever • Botulism, foodborne • Meningococcal disease • Severe acute respiratory (VHF), including Ebola virus • Cholera • Middle East Respiratory syndrome (SARS) disease, Lassa fever, Marburg • Diphtheria Syndrome (MERS) • Smallpox hemorrhagic fever, and • Influenza A – novel virus • Plague • Tularemia Crimean-Congo hemorrhagic infection • Rabies, human fever Any unexpected pattern of cases, suspected cases, deaths or increased incidence of any other disease of major public health concern, because of the severity of disease or potential for epidemic spread, which may indicate a newly recognized infectious agent, outbreak, epidemic, related public health hazard or act of bioterrorism. Class B: Disease of public health concern needing timely response because of potential for epidemic spread – report by the end of the next business day after the existence of a case, a suspected case, or a positive laboratory result is known. • Amebiasis • Carbapenemase-producing • Hepatitis B (perinatal) • Salmonellosis • Arboviral neuroinvasive and carbapenem-resistant • Hepatitis C (non-perinatal) • Shigellosis -

Diarrheal Illness
Diarrheal Illness [Announcer] This program is presented by the Centers for Disease Control and Prevention. [Karen Hunter] Hi, I’m Karen Hunter and today I’m talking with Dr. Steve Monroe, director of CDC’s Division of High-Consequence Pathogens and Pathology. Our conversation is based on his paper about viral gastroenteritis, which appears in CDC's journal, Emerging Infectious Diseases. Welcome Dr. Monroe. [Steve Monroe] Thank you Karen, it’s a pleasure to be here. [Karen Hunter] Dr. Monroe, what is viral gastroenteritis? [Steve Monroe] Gastroenteritis is an irritation of the stomach or intestinal tract. Most people experience this as severe diarrhea, vomiting, and stomach pain. For this reason, it is often referred to as stomach flu, even though it is not caused by a flu virus. The more general term is “diarrheal illness.” When caused by a virus, it is known as viral gastroenteritis. There are several viruses that can cause this illness. [Karen Hunter] Your paper focuses on two of these viruses – norovirus and rotavirus. What are the main differences between the two of them? [Steve Monroe] The main differences between norovirus and rotavirus are in the age of people most affected and in the approaches we use for control and prevention. Norovirus can infect people of all ages, while rotavirus is most commonly found in young children. And, while there’s an effective vaccine to prevent rotavirus infection, current efforts to control norovirus illness rely primarily on emphasizing good personal hygiene and infection control practices. [Karen Hunter] We’d like to hear about both of these viruses. -

Hepatitis E Virus Genotype 3 in Shellfish, United Kingdom
LETTERS term to dramatically reduce the high 3. Biswas PK, Christensen JP, Ahmed SS, biologically relevant sentinels and incidence of HPAI in Bangladesh. We Barua H, Das A, Rahman MH. at al. Avian can indicate potential pathogens that infl uenza outbreaks in chickens, Bangla- have progressively and dramatically desh. Emerg Infect Dis. 2008;14:1909–12. are contaminating the environment. increased the scope and benefi ts of our http://dx.doi.org/10.3201/eid1412.071567 It is essential to ensure that this pilot PVC implementation program, 4. Biswas PK, Christensen JP, Ahmed SS, sustainable resource of coastal areas, but additional work is needed. To Das A, Rahman MH, Barua H, et al. where mussels and oysters are farmed Risk for infection with highly pathogenic help spread PVC approaches through- avian infl uenza virus (H5N1) in back- or collected wild, is not subjected to out the country, community leaders, yard chickens, Bangladesh. Emerg In- environmental contamination that imams of local mosques, and school fect Dis. 2009;15:1931–6. http://dx.doi. could lead to public health risks. teachers can be trained to implement org/10.3201/eid1512.090643 Risk management for bivalve 5. Biswas PK, Rahman MH, Das A, Ahmed awareness programs on safe practices SS, Giasuddin M, Christensen JP. Risk mollusks, aimed at control of fecal for raising poultry and regular clean- for highly pathogenic avian infl uenza pollution, relies heavily on the use ing and disinfection of live bird mar- H5N1 virus infection in chickens in of Escherichia coli as an indicator kets. The strengthening of biosecurity small-scale commercial farms, in a high- of fecal (sewage) contamination risk area, Bangladesh, 2008. -

Emerging Water-Borne Pathogens
Appl Microbiol Biotechnol (2003) 61:424–428 DOI 10.1007/s00253-003-1302-y MINI-REVIEW S. Sharma · P. Sachdeva · J. S. Virdi Emerging water-borne pathogens Received: 1 October 2002 / Revised: 27 February 2003 / Accepted: 28 February 2003 / Published online: 9 April 2003 Springer-Verlag 2003 Abstract Emerging water-borne pathogens constitute a number of reviews and other publications available on the major health hazard in both developed and developing subject discuss emerging and re-emerging pathogens in nations. A new dimension to the global epidemiology of general, including water-borne, without addressing the cholera—an ancient scourge—was provided by the issues specifically pertinent to emerging water-borne emergence of Vibrio cholerae O139. Also, water-borne pathogens. This review critically examines which organ- enterohaemorrhagic Escherichia coli (E. coli O157:H7), isms really qualify as emerging water-borne pathogens, although regarded as a problem of the industrialized west, the possible reasons underlying their emergence and has recently caused outbreaks in Africa. Outbreaks of specific measures to face the challenge posed by them. chlorine-resistant Cryptosporidium have motivated water Due to the paucity of data, it may be difficult to decide authorities to reassess the adequacy of current water- which of all water-borne pathogens are emerging. Nev- quality regulations. Of late, a host of other organisms, ertheless, there are some clear-cut candidates. such as hepatitis viruses (including hepatitis E virus), No organism other than Vibrio cholerae could serve as Campylobacter jejuni, microsporidia, cyclospora, Yersin- a better example of an emerging water-borne pathogen. ia enterocolitica, calciviruses and environmental bacteria Cholera is an ancient scourge and to date seven like Mycobacterium spp, aeromonads, Legionella pneu- pandemics have been recorded. -

Norovirus-Gen508.Pdf
Norovirus Gastroenteritis: Management of Outbreaks in Healthcare Settings U.S. Department of Health and Human Services Centers for Disease Control and Prevention Norovirus The most common cause of cases of acute gastroenteritis and gastroenteritis outbreaks Can affect nearly everyone in the population (from children to the elderly and everyone in between!) particularly because there is no long term immunity to the virus Causes acute but self-limited diarrhea, often with vomiting, abdominal cramping, fever, and fatigue . Most individuals recover from acute symptoms with 2-3 days , but can be more severe in vulnerable populations Burden of Norovirus Infection #1 cause of acute gastroenteritis in U.S. 21 million cases annually . 1 in 14 Americans become ill each year . 71,000 hospitalized annually in U.S. 80 deaths annually among elderly in U.K. 91,000 emergency room visits overall in the U.S. Occurs year round with peak activity during the winter months Cases occur in all settings, across the globe Scallan et al. 2011. EID. 17(1): 7-15.; Patel et al. 2008. EID. 14(8); 1224-31.; Harris et al. 2008. EID. 14(10); 1546-52. Norovirus in Healthcare Facilities Norovirus is a recognized cause of gastroenteritis outbreaks in institutions. Healthcare facilities are the most commonly reported settings of norovirus gastroenteritis outbreaks in the US and other industrialized countries. Outbreaks of gastroenteritis in healthcare settings pose a risk to patients, healthcare personnel, and to the efficient provision of healthcare services. Norovirus Activity in Healthcare Incidence of norovirus outbreaks in acute care facilities and community hospitals within the United States remains unclear. -

Disease Reporting-Clinician
Infections, diseases and conditions reportable by clinicians: 2015 Report immediately • Anthrax (Bacillus anthracis) • Botulism (Clostridium botulinum) • Cholera (Vibrio cholerae O1, O139, or toxigenic) • Diphtheria (Corynebacterium diphtheriae) • Hemorrhagic fever caused by viruses of the filovirus (e.g., Ebola, Marburg) or arenavirus (e.g., Lassa, Machupo) families • Influenza (novel)1 (From footnote: Influenza A virus that cannot be subtyped by commercially distributed assays) • Marine intoxication (intoxication caused by marine microorganisms or their byproducts (e.g., paralytic shellfish poisoning, domoic acid intoxication, ciguatera, scombroid) • Measles (rubeola) • Plague (Yersinia pestis) • Poliomyelitis • Rabies (human) • Rubella • SARS (Severe Acute Respiratory Syndrome or SARS-coronavirus) • Smallpox (variola) • Tularemia (Francisella tularensis) • Yellow fever • Outbreaks and uncommon illnesses (any known or suspected common-source outbreak; any uncommon illness of potential public health significance) Report within 24 hours (including weekends and holidays) • Haemophilus influenzae (any isolation or identification from a normally sterile specimen type) • Neisseria meningitidis • Pesticide poisoning Public Health Division | Infections, diseases and conditions reportable by clinicians 128 Report within one working day • Amebic infections (central nervous system only) (for example infection by Naegleria or Balamuthia spp. • Animal bites (of humans) • Arthropod vector-borne disease (babesiosis, California encephalitis, Colorado -

What Is Norovirus?
What is norovirus? Norovirus is a serious gastrointestinal illness that causes inflammation of the stomach and/or intestines. This inflammation leads to nausea, vomiting, diarrhea, and abdominal pain. Norovirus is extremely contagious (easy to spread) from one person to another. Norovirus is not related to the flu (influenza), even though it is sometimes called the stomach flu. Anyone can get norovirus, and they can have the illness multiple times during their lifetime. Norovirus causes approximately 21 million illnesses each year. It is the leading cause of illness and outbreaks related to food in the United States. Symptoms start between 12 to 48 hours after being exposed and can last anywhere from one to three days. Symptoms include diarrhea, nausea, vomiting, and/or stomach pain. Dehydration is a big concern for people with norovirus, especially in the elderly and the very young, and a major reason for people being hospitalized. People are most contagious when they are actively sick and for the first few days after getting over the illness. How serious is norovirus? Norovirus is a serious illness that makes people feel extremely ill and vomit or have diarrhea. Most people get better within one to two days. Norovirus can be very serious among young children, the elderly, and people with other illnesses, and can lead to severe dehydration, hospitalization, and even death. How does norovirus spread? It generally spreads when infected food service workers touch food without washing their hands well or at all. Norovirus spreads from: • Person-to-person (e.g., shaking hands, sharing food or eating from the same utensils, or caring for someone who is ill with norovirus). -

Latest Ncbi-Taxonomist Docker Image Can Be Pulled from Registry.Gitlab.Com/Janpb/ Ncbi-Taxonomist:Latest
ncbi-taxonomist Documentation Release 1.2.1+8580b9b Jan P Buchmann 2020-11-15 Contents: 1 Installation 3 2 Basic functions 5 3 Cookbook 35 4 Container 39 5 Frequently Asked Questions 49 6 Module references 51 7 Synopsis 63 8 Requirements and Dependencies 65 9 Contact 67 10 Indices and tables 69 Python Module Index 71 Index 73 i ii ncbi-taxonomist Documentation, Release 1.2.1+8580b9b 1.2.1+8580b9b :: 2020-11-15 Contents: 1 ncbi-taxonomist Documentation, Release 1.2.1+8580b9b 2 Contents: CHAPTER 1 Installation Content • Local pip install (no root required) • Global pip install (root required) ncbi-taxonomist is available on PyPi via pip. If you use another Python package manager than pip, please consult its documentation. If you are installing ncbi-taxonomist on a non-Linux system, consider the propsed methods as guidelines and adjust as required. Important: Please note If some of the proposed commands are unfamiliar to you, don’t just invoke them but look them up, e.g. in man pages or search online. Should you be unfamiliar with pip, check pip -h Note: Python 3 vs. Python 2 Due to co-existing Python 2 and Python 3, some installation commands may be invoked slighty different. In addition, development and support for Python 2 did stop January 2020 and should not be used anymore. ncbi-taxonomist requires Python >= 3.8. Depending on your OS and/or distribution, the default pip command can install either Python 2 or Python 3 packages. Make sure you use pip for Python 3, e.g.