Hepatitis E Virus
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Towards the Improved Accuracy of Hepatitis E Diagnosis In
healthcare Review Towards the Improved Accuracy of Hepatitis E Diagnosis in Vulnerable and Target Groups: A Global Perspective on the Current State of Knowledge and the Implications for Practice Jasminka Talapko 1 , Tomislav Meštrovi´c 2,3, Emina Pustijanac 4 and Ivana Škrlec 1,* 1 Faculty of Dental Medicine and Health, Josip Juraj Strossmayer University of Osijek, HR-31000 Osijek, Croatia; [email protected] 2 University Centre Varaždin, University North, HR-42000 Varaždin, Croatia; [email protected] 3 Clinical Microbiology and Parasitology Unit, Dr. Zora Profozi´cPolyclinic, HR-10000 Zagreb, Croatia 4 Faculty of Natural Sciences, Juraj Dobrila University of Pula, HR-52100 Pula, Croatia; [email protected] * Correspondence: [email protected] Abstract: The hepatitis E virus (HEV) is a positive single-stranded, icosahedral, quasi-enveloped RNA virus in the genus Orthohepevirus of the family Hepeviridae. Orthohepevirus A is the most numerous species of the genus Orthohepevirus and consists of eight different HEV genotypes that can cause infection in humans. HEV is a pathogen transmitted via the fecal–oral route, most commonly by consuming fecally contaminated water. A particular danger is the HEV-1 genotype, which poses a very high risk of vertical transmission from the mother to the fetus. Several outbreaks caused by this genotype have been reported, resulting in many premature births, abortions, and also neonatal Citation: Talapko, J.; Meštrovi´c,T.; and maternal deaths. Genotype 3 is more prevalent in Europe; however, due to the openness of Pustijanac, E.; Škrlec, I. Towards the the market, i.e., trade-in animals which represent a natural reservoir of HEV (such as pigs), there is Improved Accuracy of Hepatitis E a possibility of spreading HEV infections outside endemic areas. -
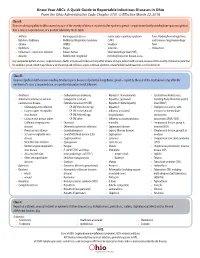
The Know Your Abcs: a Quick Guide to Reportable Infectious Diseases
Know Your ABCs: A Quick Guide to Reportable Infectious Diseases in Ohio From the Ohio Administrative Code Chapter 3701-3; Effective March 22, 2018 Class A: Diseases of major public health concern because of the severity of disease or potential for epidemic spread – report immediately via telephone upon recognition that a case, a suspected case, or a positive laboratory result exists. • Anthrax • Meningococcal disease • Severe acute respiratory syndrome fever, Marburg hemorrhagic fever, • Botulism, foodborne • Middle East Respiratory Syndrome (SARS) and Crimean-Congo hemorrhagic • Cholera (MERS) • Smallpox fever • Diphtheria • Plague • Tularemia • Yellow fever • Influenza A – novel virus infection • Rabies, human • Viral hemorrhagic fever (VHF), • Measles • Rubella (not congenital) including Ebola virus disease, Lassa Any unexpected pattern of cases, suspected cases, deaths or increased incidence of any other disease of major public health concern, because of the severity of disease or potential for epidemic spread, which may indicate a newly recognized infectious agent, outbreak, epidemic, related public health hazard or act of bioterrorism. Class B: Disease of public health concern needing timely response because of potential for epidemic spread – report by the end of the next business day after the existence of a case, a suspected case, or a positive laboratory result is known. • Amebiasis • Carbapenemase-producing • Hepatitis C (non-perinatal) • Spotted Fever Rickettsiosis, • Arboviral neuroinvasive and non- carbapenem-resistant • Hepatitis C (perinatal) including Rocky Mountain spotted neuroinvasive disease: Enterobacteriaceae (CP-CRE) • Hepatitis D (delta hepatitis) fever (RMSF) • Chikungunya virus infection • CP-CRE Enterobacter spp. • Hepatitis E • Staphylococcus aureus, with • Eastern equine encephalitis • CP-CRE Escherichia coli • Influenza-associated resistance or intermediate virus disease • CP-CRE Klebsiella spp. -

Reportable Diseases and Conditions
KINGS COUNTY DEPARTMENT of PUBLIC HEALTH 330 CAMPUS DRIVE, HANFORD, CA 93230 REPORTABLE DISEASES AND CONDITIONS Title 17, California Code of Regulations, §2500, requires that known or suspected cases of any of the diseases or conditions listed below are to be reported to the local health jurisdiction within the specified time frame: REPORT IMMEDIATELY BY PHONE During Business Hours: (559) 852-2579 After Hours: (559) 852-2720 for Immediate Reportable Disease and Conditions Anthrax Escherichia coli: Shiga Toxin producing (STEC), Rabies (Specify Human or Animal) Botulism (Specify Infant, Foodborne, Wound, Other) including E. coli O157:H7 Scrombroid Fish Poisoning Brucellosis, Human Flavivirus Infection of Undetermined Species Shiga Toxin (Detected in Feces) Cholera Foodborne Disease (2 or More Cases) Smallpox (Variola) Ciguatera Fish Poisoning Hemolytic Uremic Syndrome Tularemia, human Dengue Virus Infection Influenza, Novel Strains, Human Viral Hemorrhagic Fever (Crimean-Congo, Ebola, Diphtheria Measles (Rubeola) Lassa, and Marburg Viruses) Domonic Acid Poisoning (Amnesic Shellfish Meningococcal Infections Yellow Fever Poisoning) Novel Virus Infection with Pandemic Potential Zika Virus Infection Paralytic Shellfish Poisoning Plague (Specify Human or Animal) Immediately report the occurrence of any unusual disease OR outbreaks of any disease. REPORT BY PHONE, FAX, MAIL WITHIN ONE (1) WORKING DAY Phone: (559) 852-2579 Fax: (559) 589-0482 Mail: 330 Campus Drive, Hanford 93230 Conditions may also be reported electronically via the California -
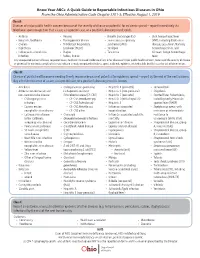
Know Your Abcs: a Quick Guide to Reportable Infectious Diseases in Ohio
Know Your ABCs: A Quick Guide to Reportable Infectious Diseases in Ohio From the Ohio Administrative Code Chapter 3701-3; Effective August 1, 2019 Class A: Diseases of major public health concern because of the severity of disease or potential for epidemic spread – report immediately via telephone upon recognition that a case, a suspected case, or a positive laboratory result exists. • Anthrax • Measles • Rubella (not congenital) • Viral hemorrhagic fever • Botulism, foodborne • Meningococcal disease • Severe acute respiratory (VHF), including Ebola virus • Cholera • Middle East Respiratory syndrome (SARS) disease, Lassa fever, Marburg • Diphtheria Syndrome (MERS) • Smallpox hemorrhagic fever, and • Influenza A – novel virus • Plague • Tularemia Crimean-Congo hemorrhagic infection • Rabies, human fever Any unexpected pattern of cases, suspected cases, deaths or increased incidence of any other disease of major public health concern, because of the severity of disease or potential for epidemic spread, which may indicate a newly recognized infectious agent, outbreak, epidemic, related public health hazard or act of bioterrorism. Class B: Disease of public health concern needing timely response because of potential for epidemic spread – report by the end of the next business day after the existence of a case, a suspected case, or a positive laboratory result is known. • Amebiasis • Carbapenemase-producing • Hepatitis B (perinatal) • Salmonellosis • Arboviral neuroinvasive and carbapenem-resistant • Hepatitis C (non-perinatal) • Shigellosis -

Seroprevalence of Anti-Hepatitis E Virus Antibodies in Domestic Pigs In
García-Hernández et al. BMC Veterinary Research (2017) 13:289 DOI 10.1186/s12917-017-1208-z RESEARCH ARTICLE Open Access Seroprevalence of anti-hepatitis E virus antibodies in domestic pigs in Mexico Montserrat Elemi García-Hernández1, Mayra Cruz-Rivera2, José Iván Sánchez-Betancourt1, Oscar Rico-Chávez1, Arely Vergara-Castañeda3, María E. Trujillo1 and Rosa Elena Sarmiento-Silva1* Abstract Background: Hepatitis E virus (HEV) infection is one of the most common causes of acute liver diseases in humans worldwide. In developing countries, HEV is commonly associated with waterborne outbreaks. Conversely, in industrialized countries, HEV infection is often associated with travel to endemic regions or ingestion of contaminated animal products. Limited information on both, human and animal HEV infection in Mexico is available. As a consequence, the distribution of the virus in the country is largely unknown. Here, we assessed the seroprevalence of HEV among swine in different geographical regions in Mexico. Methods: Seroprevalence of anti-HEV antibodies in swine herds in Mexico was evaluated in a representative sample including 945 pig serum specimens from different regions of the country using a commercial enzyme-linked immunosorbent assay (ELISA). Results: The overall prevalence of anti-HEV antibodies in swine was 59.4%. The northern region of Mexico exhibited the highest seroprevalence in the country (86.6%), while the central and southern regions in Mexico showed lower seroprevalence, 42.7% and 51.5%, respectively. Conclusions: In Mexico, HEV seroprevalence in swine is high. Importantly, northern Mexico showed the highest seroprevalence in the country. Thus, further studies are required to identify the risk factors contributing to HEV transmission among pigs in the country. -

Characterizing and Evaluating the Zoonotic Potential of Novel Viruses Discovered in Vampire Bats
viruses Article Characterizing and Evaluating the Zoonotic Potential of Novel Viruses Discovered in Vampire Bats Laura M. Bergner 1,2,* , Nardus Mollentze 1,2 , Richard J. Orton 2 , Carlos Tello 3,4, Alice Broos 2, Roman Biek 1 and Daniel G. Streicker 1,2 1 Institute of Biodiversity, Animal Health and Comparative Medicine, College of Medical, Veterinary and Life Sciences, University of Glasgow, Glasgow G12 8QQ, UK; [email protected] (N.M.); [email protected] (R.B.); [email protected] (D.G.S.) 2 MRC–University of Glasgow Centre for Virus Research, Glasgow G61 1QH, UK; [email protected] (R.J.O.); [email protected] (A.B.) 3 Association for the Conservation and Development of Natural Resources, Lima 15037, Peru; [email protected] 4 Yunkawasi, Lima 15049, Peru * Correspondence: [email protected] Abstract: The contemporary surge in metagenomic sequencing has transformed knowledge of viral diversity in wildlife. However, evaluating which newly discovered viruses pose sufficient risk of infecting humans to merit detailed laboratory characterization and surveillance remains largely speculative. Machine learning algorithms have been developed to address this imbalance by ranking the relative likelihood of human infection based on viral genome sequences, but are not yet routinely Citation: Bergner, L.M.; Mollentze, applied to viruses at the time of their discovery. Here, we characterized viral genomes detected N.; Orton, R.J.; Tello, C.; Broos, A.; through metagenomic sequencing of feces and saliva from common vampire bats (Desmodus rotundus) Biek, R.; Streicker, D.G. and used these data as a case study in evaluating zoonotic potential using molecular sequencing Characterizing and Evaluating the data. -

HEPATITIS E VIRCLIA® Igm MONOTEST
EN 1 N KIT FEATURES: HEPATITIS E VIRCLIA® IgM All reagents supplied are ready to use. Serum dilution solution and conjugate are coloured to help in MONOTEST the performance of the technique. For in vitro diagnostic use Sample predilution is not necessary. Reagents required for the run of the test are included in the VCM067: Indirect chemiluminescent immunoassay (CLIA) to monodose presentation. test IgM antibodies against hepatitis E virus in human serum/plasma. 24 tests. KIT CONTENTS: 1 VIRCLIA® HEPATITIS E IgM MONODOSE: 24 monodoses consisting of 3 reaction wells and 5 reagent wells with de INTRODUCTION: following composition : Hepatitis E virus (HEV) is the causative agent of hepatitis E. HEV Wells A, B, C: reaction wells; wells coated with anti-IgM belongs to the family Hepeviridae. Four out of seven genotypes antibodies (µ-specific). of Orthohepevirus A are known to infect humans. Genotypes 1 Well D: Conjugate: orange; containing HEV recombinant and 2 are responsible for human infections exclusively and are antigen, peroxidase conjugate dilution and Neolone and endemic in Asia, Africa and some American countries. Bronidox as preservatives. Genotypes 3 and 4 are zoonotic and present in Europe, United Well E: Serum dilution solution: blue; phosphate buffer States, Japan, China and Taiwan. HEV is transmitted via the fecal-oral route. Foodborne infection can occur from containing protein stabilizers and Neolone and Bronidox as consumption of uncooked/undercooked meat or organs from preservatives. infected animals. A wide range of clinical manifestations, from Well F: Calibrator: clear; positiveONLY serum dilution containing asymptomatic or subclinical to acute liver failure, can be Neolone and Bronidox as preservatives. -

Hepatitis E Virus Genotype 3 in Shellfish, United Kingdom
LETTERS term to dramatically reduce the high 3. Biswas PK, Christensen JP, Ahmed SS, biologically relevant sentinels and incidence of HPAI in Bangladesh. We Barua H, Das A, Rahman MH. at al. Avian can indicate potential pathogens that infl uenza outbreaks in chickens, Bangla- have progressively and dramatically desh. Emerg Infect Dis. 2008;14:1909–12. are contaminating the environment. increased the scope and benefi ts of our http://dx.doi.org/10.3201/eid1412.071567 It is essential to ensure that this pilot PVC implementation program, 4. Biswas PK, Christensen JP, Ahmed SS, sustainable resource of coastal areas, but additional work is needed. To Das A, Rahman MH, Barua H, et al. where mussels and oysters are farmed Risk for infection with highly pathogenic help spread PVC approaches through- avian infl uenza virus (H5N1) in back- or collected wild, is not subjected to out the country, community leaders, yard chickens, Bangladesh. Emerg In- environmental contamination that imams of local mosques, and school fect Dis. 2009;15:1931–6. http://dx.doi. could lead to public health risks. teachers can be trained to implement org/10.3201/eid1512.090643 Risk management for bivalve 5. Biswas PK, Rahman MH, Das A, Ahmed awareness programs on safe practices SS, Giasuddin M, Christensen JP. Risk mollusks, aimed at control of fecal for raising poultry and regular clean- for highly pathogenic avian infl uenza pollution, relies heavily on the use ing and disinfection of live bird mar- H5N1 virus infection in chickens in of Escherichia coli as an indicator kets. The strengthening of biosecurity small-scale commercial farms, in a high- of fecal (sewage) contamination risk area, Bangladesh, 2008. -

Emerging Water-Borne Pathogens
Appl Microbiol Biotechnol (2003) 61:424–428 DOI 10.1007/s00253-003-1302-y MINI-REVIEW S. Sharma · P. Sachdeva · J. S. Virdi Emerging water-borne pathogens Received: 1 October 2002 / Revised: 27 February 2003 / Accepted: 28 February 2003 / Published online: 9 April 2003 Springer-Verlag 2003 Abstract Emerging water-borne pathogens constitute a number of reviews and other publications available on the major health hazard in both developed and developing subject discuss emerging and re-emerging pathogens in nations. A new dimension to the global epidemiology of general, including water-borne, without addressing the cholera—an ancient scourge—was provided by the issues specifically pertinent to emerging water-borne emergence of Vibrio cholerae O139. Also, water-borne pathogens. This review critically examines which organ- enterohaemorrhagic Escherichia coli (E. coli O157:H7), isms really qualify as emerging water-borne pathogens, although regarded as a problem of the industrialized west, the possible reasons underlying their emergence and has recently caused outbreaks in Africa. Outbreaks of specific measures to face the challenge posed by them. chlorine-resistant Cryptosporidium have motivated water Due to the paucity of data, it may be difficult to decide authorities to reassess the adequacy of current water- which of all water-borne pathogens are emerging. Nev- quality regulations. Of late, a host of other organisms, ertheless, there are some clear-cut candidates. such as hepatitis viruses (including hepatitis E virus), No organism other than Vibrio cholerae could serve as Campylobacter jejuni, microsporidia, cyclospora, Yersin- a better example of an emerging water-borne pathogen. ia enterocolitica, calciviruses and environmental bacteria Cholera is an ancient scourge and to date seven like Mycobacterium spp, aeromonads, Legionella pneu- pandemics have been recorded. -

Classification and Genomic Diversity of Enterically Transmitted Hepatitis Viruses
Downloaded from http://perspectivesinmedicine.cshlp.org/ on October 2, 2021 - Published by Cold Spring Harbor Laboratory Press Classification and Genomic Diversity of Enterically Transmitted Hepatitis Viruses Donald B. Smith1,2 and Peter Simmonds2 1Centre for Immunity, Infection and Evolution, University of Edinburgh, Edinburgh EH9 3JT, United Kingdom 2Nuffield Department of Medicine, University of Oxford, Oxford OX1 3SY, United Kingdom Correspondence: [email protected] Hepatitis A virus (HAV) and hepatitis E virus (HEV) are significant human pathogens and are responsible for a substantial proportion of cases of severe acute hepatitis worldwide. Genetically, both viruses are heterogeneous and are classified into several genotypes that differ in their geographical distribution and risk group association. There is, however, little evidence that variants of HAVor HEV differ antigenically or in their propensity to cause severe disease. Genetically more divergent but primarily hepatotropic variants of both HAVand HEV have been found in several mammalian species, those of HAV being classified into eight species within the genus Hepatovirus in the virus family Picornaviridae. HEV is classified as a member of the species Orthohepevirus A in the virus family Hepeviridae, a species that additionally contains viruses infecting pigs, rabbits, and a variety of other mammalian species. Other species (Orthohepevirus B–D) infect a wide range of other mammalian species including rodents and bats. epatitis Avirus (HAV) and hepatitis E virus ease in humans. However, the two viruses are H(HEV) show moderate genetic diversity, quite distinct in terms of their replication strat- both being classified into several genotypes in- egy, virion structure, and taxonomy (see Kenney fecting humans, and several additional species and Meng 2018; McKnight and Lemon 2018). -

Disease Reporting-Clinician
Infections, diseases and conditions reportable by clinicians: 2015 Report immediately • Anthrax (Bacillus anthracis) • Botulism (Clostridium botulinum) • Cholera (Vibrio cholerae O1, O139, or toxigenic) • Diphtheria (Corynebacterium diphtheriae) • Hemorrhagic fever caused by viruses of the filovirus (e.g., Ebola, Marburg) or arenavirus (e.g., Lassa, Machupo) families • Influenza (novel)1 (From footnote: Influenza A virus that cannot be subtyped by commercially distributed assays) • Marine intoxication (intoxication caused by marine microorganisms or their byproducts (e.g., paralytic shellfish poisoning, domoic acid intoxication, ciguatera, scombroid) • Measles (rubeola) • Plague (Yersinia pestis) • Poliomyelitis • Rabies (human) • Rubella • SARS (Severe Acute Respiratory Syndrome or SARS-coronavirus) • Smallpox (variola) • Tularemia (Francisella tularensis) • Yellow fever • Outbreaks and uncommon illnesses (any known or suspected common-source outbreak; any uncommon illness of potential public health significance) Report within 24 hours (including weekends and holidays) • Haemophilus influenzae (any isolation or identification from a normally sterile specimen type) • Neisseria meningitidis • Pesticide poisoning Public Health Division | Infections, diseases and conditions reportable by clinicians 128 Report within one working day • Amebic infections (central nervous system only) (for example infection by Naegleria or Balamuthia spp. • Animal bites (of humans) • Arthropod vector-borne disease (babesiosis, California encephalitis, Colorado -

Infant Botulism ESCMID Online Lecture Library
Emerging issues in food-borne infection ‘We never look for it because we never find it’ – emerging pathogens we need to know about ©John by Threlfall author ESCMID Online UKLecture Library ECCMID April 2013 ‘We never look for it because we never find it’ – emerging pathogens we need to know about ‘There are known knowns; there are things we know we know. We also know there are known unknowns; that is to say, we know there are some things we do not know’. ‘But there are also unknown unknowns – the ones we don’t know we don’t know’.© by author ESCMID Online Lecture Library Donald Rumsfeld, US Defence Secretary, 2002 ‘We never look for it because we never find it’ - emerging pathogens we need to know about Questions (1) 1. What is the trigger for recognising the emergence of a ‘new’ pathogen? Possible answers: Increase in incidence above ‘expected’ norm. Outbreak of ‘new’ strain. Increase in mortality in vulnerable population groups. 2. Which ‘emerging pathogen’© by hasauthor been responsible for major outbreak(s) of food-poisoning in Europe over the last five years, involving numerous fatalities? ESCMID Online Lecture Library Possible answers: Norovirus Cryptosporidium Hepatitis E (HEV) Escherichia coli O104:H4 ‘We never look for it because we never find it’ – emerging pathogens we need to know about Disease / Organism • What are the disease symptoms? • What is the causative organism? • How rapidly is this isolated? • How common is the organism? – individuals, communities, environment Source / Reservoir © by author • Is it a food contamination issue? • ESCMIDIf so, what is the food? Online Lecture Library • What is the food animal reservoir (if any?) ‘We never look for it because we never find it’ - emerging pathogens we need to know about Questions (2) 1.